January 15, 2009 (Vol. 29, No. 2)
Technology Useful for High-Throughput Screening and Lead-Optimization Projects
Approximately 40% of today’s pharmaceuticals act on proteins such as GPCRs and ion channels that are embedded within cell membranes. These transmembrane proteins form the functional environmental interface of the cell, accounting for their value as therapeutic targets. They have been exploited to treat a wide range of diseases, including cancer, heart disease, diabetes, nervous system disorders, and infectious diseases.
Studying membrane protein structure and interactions during drug characterization and optimization, however, presents a unique set of challenges. Typically, membrane protein structural conformation is dependent on the presence of the cellular lipid membrane. Conventional binding assays for membrane proteins employ live cells or membrane preparations derived from cells to ensure structural stability of the embedded target proteins.
These formats, however, exhibit low target protein concentration, receptor heterogeneity, lipid contaminants, and, in the case of membrane preparations, receptor inversion, leading to poor sensitivity and high experimental variability. Furthermore, because of their large size and heterogeneity, cells and membrane vesicles are not amenable to advanced detection devices that employ microfluidic systems such as optical biosensors.
A number of novel strategies have attempted to isolate membrane proteins from the lipid bilayer of the cell and present them in a high concentration, homogeneous format. Most techniques involve the reconstitution of receptors onto artificial structures, such as beads or micelles. These methods use detergents to control protein unfolding and micelle incorporation. Since detergents and artificial lipids can alter membrane protein structure, conditions must be determined empirically and validated for each membrane protein studied.
An alternative to detergent-based manipulation is to express membrane proteins in biological particles, such as intracellular compartments of specialized bacteria. While bacterial systems offer the benefits of high concentration protein expression, target proteins must typically be fused with native anchor proteins to ensure trafficking to membrane surfaces. The lack of mammalian post-translational modifications and the differences in cell membrane composition can also influence protein structure and function.
Lipoparticles
A novel format for the experimental manipulation of membrane proteins, and one that overcomes the restrictions of existing approaches, is Lipoparticle technology from Integral Molecular. Lipoparticles are virus-like particles that present high concentrations of membrane protein on their surfaces. Because embedded membrane proteins are captured directly from mammalian cell surfaces, their native structure is preserved without the need for detergents or fusion partners. This technology, which can be used to isolate any plasma membrane protein, results in a nanoparticulate suspension of the target protein that can be utilized in diverse assays where the use of cells or membrane preparations is limited.
Lipoparticles are produced from mammalian cells by co-expressing the retroviral structural core polyprotein Gag, along with a desired membrane protein. Each protein traffics to the cell membrane, where Gag proteins self assemble into core capsid structures which bud through the plasma membrane, capturing target membrane proteins (Figure 1A).
Because Lipoparticle formation is an active process mediated by Gag, the resulting particles are morphologically homogeneous. Dynamic light scattering and electron microscopy confirm they are free of contaminating cellular debris (Figure 1B). With a mean diameter of approximately 150 nm and a narrow size distribution (Figure 1C), they are readily suspended in aqueous buffers.
Radioligand binding assays using Lipoparticle suspensions confirm the functional integrity of incorporated membrane proteins, as shown for Lipoparticles containing the chemokine receptor CXCR4 (Figure 2A). Binding of the CXCR4 ligand, SDF-1a, is specific, concentration-dependent, and saturable, exhibiting a low nanomolar KD. Its similarities to cell-bound receptor behavior confirm that the Lipoparticle-incorporated receptor is structurally and functionally intact.
The CXCR4 competitive binding curve indicates an estimated Lipoparticle receptor content of approximately 200 pmol/mg total protein, which is approximately 20–100 fold higher than what is found in conventional membrane preparations. Similar receptor concentrations, typically between 50–200 pmol/mg, have been determined for a diverse range of other membrane proteins incorporated into Lipoparticles.
The high concentration of membrane proteins in Lipoparticles enables their use for rapid screening of membrane protein interactions in conventional and nonconventional formats. The binding of a diverse panel of antibodies with specificity for the chemokine receptor CCR5 was assessed using ELISA in 96-well microplates. When Lipoparticles incorporating CCR5 were included in the plates, high levels of binding were observed for all antibodies, including several (45523, 45529, and 45517) that recognize complex, nonlinear epitopes (Figure 2B).
None of the antibodies recognized Lipoparticles incorporating a different chemokine receptor (CXCR4). The highly concentrated, homogeneous membrane proteins contained in Lipoparticle suspensions enable drug and antibody screening in high-throughput formats that have previously been convenient only for soluble proteins.
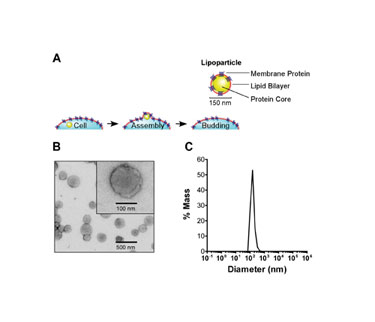
Figures: 1A, 1B and 1C
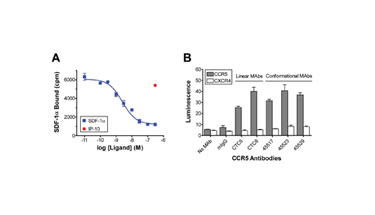
Figures: 2A and 2B
Optical Biosensors
Because Lipoparticle suspensions approximate the physical properties of membrane proteins in solution, they are ideal for studying membrane protein interactions in mobile-phase microfluidic systems such as optical biosensors. When proteins such as antibodies are immobilized to a biosensor chip, the binding of molecules that are flowed across the chip can be optically detected and quantified by changes in refractive index.
Nanosized Lipoparticles are readily flowed through the 50 µm channel of an optical biosensor, enabling their use as mobile-phase reagents, as shown here for the binding interaction between chip-immobilized antibodies and Lipoparticles containing the cannabinoid receptor (CB1). Lipoparticle binding to flow cells on which anti-CB1 antibodies were immobilized was readily detectable, while Lipoparticles did not bind to flow cells on which a nonspecific monoclonal antibody was immobilized (Figure 3).
Lipoparticles can also be immobilized directly on optical biosensor surfaces (Figure 4A). While conventional binding assays allow only for measurements of equilibrium binding affinity, immobilization to a biosensor chip permits on and off rates of the binding interaction to be measured in real time.
Lipoparticles incorporating the chemokine receptor CXCR4 were immobilized on a biosensor chip. Sensorgrams measure the concentration-dependent association of the anti-CXCR4 monoclonal antibody 12G5 as it was flowed across the chip, and its slow dissociation when buffer alone was flowed (Figure 4B). Sensorgram analyses from diverse antibodies against CXCR4 show a distinct kinetic profile for each antibody despite similar equilibrium binding affinities in a number of cases (Figure 4C).
Lipoparticles are useful for studying membrane protein interactions, enabling homogeneous suspension of purified, high-concentration receptors. The technology is compatible with numerous assay formats and is especially appropriate for high-throughput screening and lead-optimization studies. The capture of complex proteins on Lipoparticles generates new avenues for investigating membrane protein interactions using novel techniques that were previously difficult to apply.
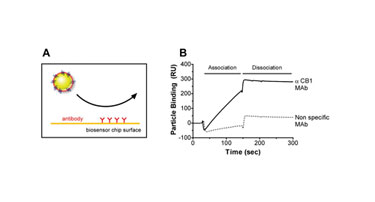
Figure 3

Figures: 4A, 4B, and 4C
Soma S.R. Banik, Ph.D., is a research scientist, Christopher J. Laing, Ph.D., is a consultant, and Benjamin J. Doranz, Ph.D. ([email protected]), is the president and CEO of Integral Molecular.



