November 15, 2006 (Vol. 26, No. 20)
Sample Preparation Strategies to Keep Pace with Faster Analytical Instruments
Most procedures for extracting DNA from general substrates involve manipulations that reduce yields and expose the sample to potential contamination. Additionally, most methods also require centrifugation, vacuum, or solvent extraction—steps that render them difficult for automation.
prepGEM™ is ZyGEM’s (www.zygem.com) closed-tube DNA-extraction procedure using a thermophilic proteinase that produces good DNA yields from a wide range of substrates. The reaction is controlled by a temperature-shift regime programmed into a thermal cycler, water-bath, or oven, eliminating the need for solvent extraction or column purification.
The new method is ideally suited to high-throughput screening applications, viral diagnostics, and forensic profiling where exposure to extraneous DNA must be prevented. In addition, the simplicity of the procedure makes it suitable for automation.
This technique has been successfully applied to a wide range of substrates. Examples are blood, tissue, and hair for high-throughput human and livestock screening for mapping and quantitative trait loci discovery programs, as well as medical samples such as tissue, blood, skin, and mucosal swabs. It has also been used in screening for environmental sample pathogen detection and speciation of insects for the Bar Code of Life Consortium project.
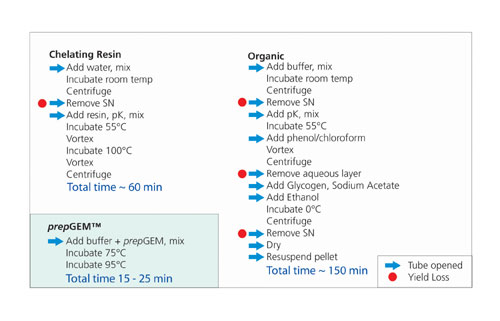
Figure 1
Keeping Pace Downstream
Many high-throughput screening regimes use blood, tissue, hair, or mucosal samples for PCR, trait, and viral diagnostic analysis. While many of these methods are highly effective at producing good-quality DNA, relatively few are easily automated, and many of the steps expose the sample to potential contamination by the technician or extraneous DNA.
prepGEM is effective for extracting DNA from blood, saliva, tissue, follicle, milk, mucosal, and bacterial swabs. The proteinase works well when DNA yields are high and dilutions are required prior to PCR amplification (as is the case with fresh blood). However, a real benefit of the method is the high-throughput automation and ability to scale down to small volumes. The simplicity and closed-tube nature of the method maximizes yield and the compatibility of the extraction buffer with PCR means that the extract can go directly into the amplification reaction.
The extraction procedure involves heating a buffered suspension of sample to 75°C for 15 min in the presence of buffer and enzyme (Figure 1). Buffer and/or sample volume can be easily altered to control yield while maintaining enzyme concentration. As little as 5-µL extraction volume can be used for just a few cells, or as much as 850 µL for a whole buccal swab has been successfully demonstrated. A subsequent incubation for 5–15 minutes at 95°C kills the enzyme, and the extract is available for immediate PCR.
The main advantage of using a proteinase that is optimally active at high temperatures is that DNA is released from the cellular material at temperatures where contaminating nucleases are inactive. Nucleases are therefore rapidly hydrolyzed by the proteinase before any degradation of the DNA occurs.
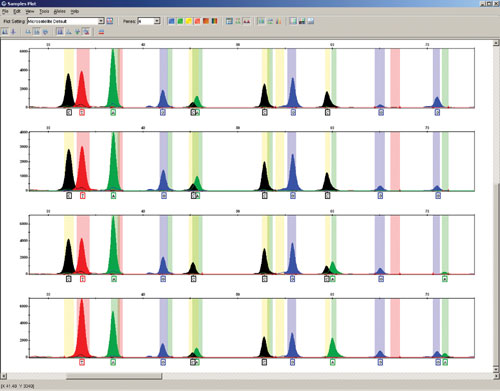
Figure 2
The Value
Comparison of the quality of STR and SNP profiles obtained from many sample types in high-throughput trait testing illustrates ZyGEM’s efficacy in comparison to currently employed solid matrix and organic solvent procedures (Figures 2 and 3). This result is significant for a number of reasons:
• Laboratories often deal with samples of marginal quantities of DNA. By removing steps in extraction methodology (pre-washes, solvent extraction, and ethanol precipitation) yields can be substantially improved.
• All procedural steps are easily automated. Any off-the-shelf liquid-handling robot coupled with a thermal cycler, water-bath, or oven can be employed for high-throughput processing.
• The extraction method is closed-tube and therefore less susceptible to contamination by extraneous DNA.
• The same extraction method is universal regardless of substrate—sample to PCR in as little as 15–25 minutes for batches of hundreds of samples.
This new method is a particularly attractive option for laboratories dealing with large volumes and those wishing to automate their DNA-extraction procedures to achieve a higher throughput and greater certainty of interpretable data for target identification and general use samples.
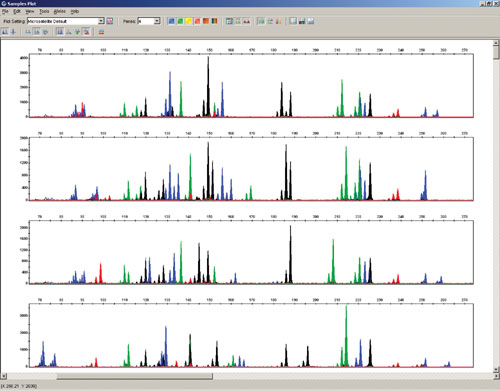
Figure 3
Geoff Corbett is senior product development manager at ZyGEM. Web: www.zygem.com. Phone: +64 7 857 0873. E-mail: [email protected].







