October 15, 2012 (Vol. 32, No. 18)
PCR continues to revolutionize molecular diagnostics. Its ability to enrich genomic regions of interest for next-generation sequencing (NGS) and interface with other technologies such as mass spectrometry are helping move the field closer to the goal of personalized medicine.
CHI’s recent “Next Generation Diagnostics Summit” highlighted a number of advances in R&D in this arena.
FFPE samples represent one of the most abundant sources of readily available specimens from which to mine genetic information. But, they also are challenging to analyze via NGS. Elizabeth Mambo, Ph.D., senior scientist, technology development, Asuragen, reported on the company’s work to address the needs for mutation detection in such technically challenging samples.
“NGS platforms are an indispensable tool for deep sequencing of patient tissue samples. But FFPE and fine needle aspiration (FNA) specimens are particularly difficult to work with due to their chemical alteration (FFPE) or limited amounts (FNA and FFPE). We developed multiple PCR-based target-enrichment methods to assess such samples and have generated data from over 170 FFPE and FNA specimens utilizing different enrichment methods and NGS platforms.”
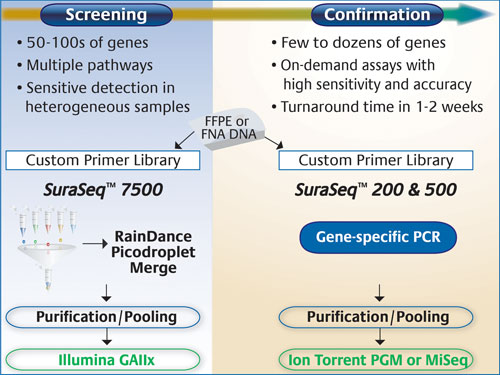
For screening purposes, the company says it utilized the RainDance RDT 1000 (RainDance Technologies), with its massively parallel picoliter droplet PCR for enrichment of up to 20,000 genomic regions, and Illumina’s Genome Analyzer that allows large-scale gene profiling or genome-wide discovery at maximal sensitivity.
For more focused screening, or confirmatory purposes, they employed the Ion Torrent PGM (Life Technologies) to achieve 3–5 million reads in a 2 hour run time. The company has also recently acquired a MiSeq (Illumina) for similar studies.
“To date, we’ve developed three PCR-based enrichment panels,” said Gary Latham, Ph.D., vp of research and technology development. “SuraSeq™ 7500, which represents more than 7,500 distinct mutations across 52 cancer genes and interrogates over 120,000 unique bases from DNA inputs as low as 250 nanograms; and SuraSeq 200 and 500, which are ideal for high-throughput, focused sequencing in commonly mutated cancer genes and require only 10–40 nanograms of DNA.”
“It is important to use a stable of instruments that rely on different sequencing chemistries,” Dr. Latham advised. “Depending on the panel size, there can be many hits, so validating positives on an orthogonal platform provides much greater confidence in the results.
“This issue is particularly critical for high-depth (>1,000x) amplicon sequencing. There are no suitable algorithms in the public domain to properly analyze such data, so we created a customized pipeline to achieve a sensitivity of 4–5% variant.”
The company will continue developing off-the-shelf and custom panels, as well as pursue implementation in its CLIA-certified laboratory.
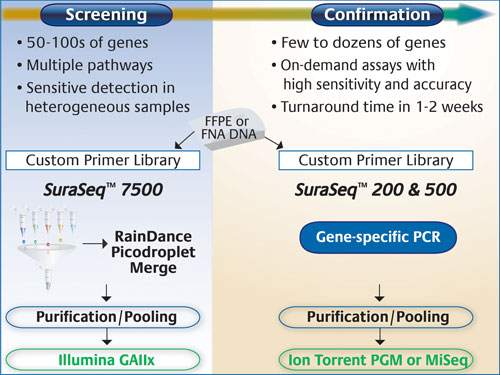
Asuragen’s SuraSeq 7500 panel targets >120,000 unique bases across 52 cancer genes, and captures >7,500 annotated mutations in the COSMIC database. Mutations are identified after sequencing on Illumina instruments and applying bioinformatic algorithms developed specifically to detect low-abundance variants in DNA from FFPE tumor biopsies. Mutation calls using this panel can be confirmed using on-demand custom panels, or off-the-shelf focused panels, such as SuraSeq 200 and 500, that are sequenced on either the Ion Torrent PGM or the Illumina MiSeq.
Clinical Microbiology
Rapid identification of infectious organisms by the clinical microbiology laboratory is critical for reducing the risk of morbidity and mortality in patients.
“Unfortunately, current methods primarily rely on the rather cumbersome and slow method of culturing specimens and using traditional phenotypic analysis for characterizing pathogens. First-generation molecular diagnostics provided greater speed, but could only assay for one or at best a few pathogens, meaning that diagnosticians often had to guess which pathogen to test for,” noted Garth D. Ehrlich, Ph.D., executive director, center for Genomic Sciences, Allegheny-Singer Research Institute.
Dr. Ehrlich says a new method coupling PCR and mass spectrometry could revolutionize the way clinical labs identify pathogens.
“We have begun using a PCR-based mass spectrometry method known as PCR-electron spray ionization, time-of-flight, mass spectrometry (PCR-ESI-TOF-MS). This technology provides unparalleled sensitivity, accuracy, and breadth within the realm of molecular diagnostics for pathogen detection. It takes advantage of the exquisite sensitivity of both PCR and mass spectrometry.
“Further, global specificity derives from the use of multiple, independent genomic amplification targets. Not only can single assays detect and speciate all members of a taxonomic domain, such as eubacteria, fungi, etc., but they can do so in hours, not days.”
The instrument used by Dr. Ehrlich and colleagues is Abbott Laboratories’ Ibis T-5000 system, predecessor to the current PLEX-ID platform that is being commercialized for clinical diagnostics. He says one doesn’t have to initially guess which pathogen(s) might be causing an infection; rather the system is capable of a broad-based identification of bacteria and fungi. His studies have evaluated samples from numerous infectious and inflammatory conditions including total joint failures, osteoarthritis, chronic nonhealing wounds, and surgical site infections, among others.
“The system can semiquantitatively identify mixed populations of microbes with a readout of genotypic analysis. There are so many advantages of this PCR-mass spec system over the rather archaic method of culturing a sample.
“Use of internal calibrations provides for relative quantitative assessments among co-infecting species. It can work even following antibiotic administration. It simultaneously detects multiple species (even biofilms) and has been shown to have superiority to all other bacterial detection methods. Also, there is no need to develop new assays for each specimen type because the technology is already established and validated.”
Although the costs per tests are not high, they do necessitate the purchase of a dedicated mass spectrometer and software package. “Despite this challenge, this new technological approach has the potential to supplant the foundation of clinical microbiology and, in so doing, save many lives.”
NGS Cancer Panels
To provide a more comprehensive genetic analysis of patient tumor tissues requires a technology that is not only suitable for assessing multiple genes but also sensitive enough to do so using minimal amounts of sample.
Jin Li, M.D., Ph.D., research director, advanced diagnostics, MolecularMD, reported on the company’s studies cross validating two commercial NGS cancer panels: Ion Torrent AmpliSeq and Illumina TruSeq. MolecularMD is a privately owned molecular diagnostics company that develops and provides assays for clinical development of targeted cancer therapy.
“As personalized medicine advances, it is important to develop increasingly sensitive clinical assays that provide a much more thorough characterization of tumors with a minimal amount of sample. We developed a cross validation strategy for validating the two commercial panels based on both panels targeting the same 46 genes and sharing similar regions of interest and sensitivities of 2–5% for minor allele detection. Also, by comparing the cancer gene panels, we were able to identify the strengths and shortcomings of each of the sequencing platforms: the Ion Torrent PGM and the Illumina MiSeq.”
Dr. Li stated that while both platforms provided solutions for fast screening of critical mutations, there were important differences. “The Ion Torrent was more resistant to low-quality FFPE DNA than the Illumina. It also had the advantage of using only 10 ng FFPE sample. Another finding was that both panels detect single base substitutions and indels, but the Illumina performed this task better on indels in the homopolymers region.”
Data analysis can be a significant challenge as well. Dr. Li suggested that a manufacturer’s software is often not sufficient for high-level analysis. “We found both panels required solid validation and enhancement by our own software to derive accurate results.”
Companion Dx Quandary
Companion diagnostics are assays that evaluate proteins or genes in order to stratify a patient population to better determine who will respond most favorably (or at all) to a therapeutic.
“This is more an FDA regulatory term since some companies are now required to provide a companion diagnostic whenever they are approved for a therapeutic. The idea is to improve patient treatment by stratifying patients into responder or nonresponder populations,” noted Richard Montagna, Ph.D., senior vp, business development, Rheonix.
Dr. Montagna said the challenge for companies is to determine when to invest the time and effort to develop the companion diagnostic for the corresponding therapeutic.
“The problem with investing the effort up front is that the therapeutic or diagnostic could fail in clinical development and then the investment has been wasted. On the other hand, delaying the diagnostic development can jeopardize timely completion of the drug trials as the therapeutic and diagnostic submissions must be FDA-reviewed simultaneously. Moreover, approval for both is required to market the drug. Companies as well as the FDA are grappling with this issue.”
In the meantime, Rheonix has developed a fully automated platform for companion diagnostics called the Rheonix CARD® system that processes a range of clinical specimens.
“There are a number of platforms in the market, but none have the versatility that we have been able to achieve for this miniaturized system. For example, many systems evaluate only microliter volumes, but sometimes this is not suitable such as for septicemia. Our microfluidic-driven system uses inexpensive plastic, disposable devices with all the required functionality for multiplexing PCR of samples with volumes from 5 microliters to 5 milliliters.”
The system is also suitable for buccal swabs, whole blood, fresh tissue, and FFPE samples. Each can be placed into the system for automatic, unattended cell lysis, DNA extraction and purification, PCR-based amplification of targets, and detection of amplicons on an integrated DNA microarray.
The company will perform its first clinical study at four sites by year-end. “We have developed a Rheonix Warfarin assay for warfarin sensitivity. Patients are given warfarin to reduce the chance of stroke-causing blood clots.
“But warfarin is metabolized differently depending on patients’ genotypes. If the drug is metabolized too rapidly, they risk throwing a clot; too slowly, and they risk bleeding out in the brain. Currently assessments are made by repetitive blood tests that take weeks or months to get the right dose. Using our CARD system, a definitive test can be done within a few hours.”
Rheonix also has programs using the CARD technology to detect the presence of single nucleotide polymorphisms associated with Plavix sensitivity and KRAS markers, among others.
Clearly, the arena of molecular diagnostics is a field on the move. Modifying and integrating technologies such as PCR-based target enrichment, NGS, and mass spectrometry are leading candidates for new synergistic approaches.

Once up to six disposable plastic Rheonix CARD (left) devices, each capable of analyzing four samples, are inserted into the EncompassMDx Workstation (right), all sample preparation, analysis, and readout of results are automatically performed.