August 1, 2010 (Vol. 30, No. 14)
System Can Also Be Used for Auto-Antibody Profiling or Screening for Clinical Studies
Functional protein arrays consist of thousands of proteins immobilized on a glass slide to facilitate the study of proteome-scale protein-interaction networks, autoantibody detection for clinical diagnosis, detection for infectious diseases, and vaccine research. Despite great promise, protein arrays have not been widely accepted because of technological hurdles including generating protein content, challenges in protein immobilization, and the need for complex robotics.
To overcome these problems, Promega offers a simple way to create medium-throughput custom protein arrays using the HaloLink™ Protein Array System. HaloLink is based on HaloTag®, a 33 kDa protein tag that forms a covalent bond with its ligand. HaloTag is multifunctional and has been used for protein expression and purification, cell-imaging, protein-immobilization, and protein-interaction analyses.
HaloLink Protein Array Systems combine protein-expression systems, a novel glass slide coating, and HaloTag technology to simplify the process of making custom protein arrays. This tutorial will focus on cell-free protein-expression systems for generating protein content, but HaloLink arrays have been successfully validated using cell-based expression systems like mammalian, E.coli, and insect cells.
Cell-free protein-expression systems based on coupled transcription-translation express proteins directly from DNA templates, are easy to use, rapid, and ideal for high-throughput formats. In addition, they can be used to express toxic, membrane, and insoluble proteins and allow insertion of non-natural amino acids containing fluorescent or other reactive labels.
The HaloLink Protein Array System uses polyethylene glycol (PEG)-coated glass slides for protein capture. PEG-based hydrogels resist nonspecific adsorption of unwanted protein and minimize surface-induced denaturation of immobilized proteins. PEG-coated glass slides activated with HaloTag ligands (HaloLink slides) are used to capture proteins-of-interest (POI) fused to HaloTag that are expressed in cell-free protein-expression systems.
Attachment through HaloTag is covalent, and POI are oriented for maximum functional activity, but more importantly, no prior protein purification is necessary. Although, other protein tags like polyhisitidine and GST can be used for orienting proteins at the surface, leaching due to the reversible nature of binding limits their utility. Finally, a silicone gasket that can be attached to the slide creates 50 wells on the glass slides allowing multiple assays to be performed manually on the same slide without any specialized equipment.
The combination of cell-free expression systems and HaloTag attachment chemistry allows users to create a medium-throughput custom array and perform downstream protein-interaction applications in less than eight hours (Figure 1A).
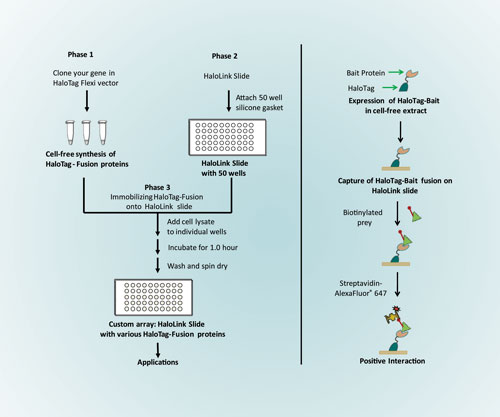
Figure 1. [left] Workflow for custom array fabrication using the HaloLink Protein Array System; [right] Strategy for protein-protein interactions using HaloLink
Detecting Protein-Protein Interactions
To demonstrate the utility of HaloLink Protein Array Systems for multiplexed protein-protein interaction studies we used five well-known protein-protein interaction pairs: p65 and p50; p65 and IκB; cJun and cFos; TAg and p53; and R1a and protein kinase A (PKA). Bait proteins (p65, cJun, TAg, and R1α) fused with HaloTag along with the prey proteins (p50, IκB, cFos, p53, and PKA) were expressed in the cell-free TNT® SP6 High-Yield Protein Expression System (Promega).
The prey proteins were labeled in vitro with biotin by the addition of Transcend™ —a protein labeling reagent (Promega) that incorporates biotinylated lysine into nascent proteins during translation.
A schematic of the protein-protein interaction assay is shown in Figure 1B. In this experiment, cell extract containing HaloTag-bait fusion proteins were added in triplicate to each well (5.0 µL/well) on a HaloLink slide and incubated for 1.0 hour to capture HaloTag-bait fusions. Immobilized bait proteins were incubated with respective biotinylated prey proteins and positive interaction was detected using AlexaFluor™ 647-labeled streptavidin.
For negative controls, preys were incubated in wells containing only HaloTag protein. The scanned slide image (Figure 2A) shows specific binding for all five protein-interaction pairs with an excellent signal-to-background ratio (Figure 2B). The ability to label proteins in vitro in a cell-free translation system in combination with the convenience of performing bait-prey interaction studies directly from lysates, significantly simplifies proteome-scale studies of protein-interaction networks.
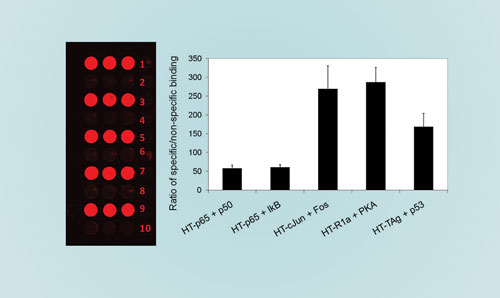
Figure 2. [left] Fluorescence scan indicating specific bait-prey protein interactions: (1) HaloTag-p65 + p50, (2) HaloTag + p50, (3) HaloTag-p65 + IkappaB, (4) HaloTag + IkappaB, (5) HaloTag-cJun + cFos, (6) HaloTag + p50, (7) HaloTag-R1alpha + PKA, (8) HaloTag + PKA, (9) HaloTag-TAg + p53, and (10) HaloTag + p53. [right] Ratio of specific to nonspecific signal was calculated for each bait-prey interacting pair. [Reprinted with permission from the Journal of Proteome Research]
Detecting Protein-DNA Interactions
The HaloLink Protein Array System can also be used for protein-nucleic acid interaction studies. Traditionally these interactions are detected using electophoretic mobility shift and fluorescence anisotropy assays. These methods need purified proteins, are labor intensive, and have low throughput. HaloLink, in contrast, is rapid, requires no purified protein, and provides a medium-throughput format.
To show that HaloLink can be used to study protein-nucleic acid interactions we selected: interaction between DNA binding protein p50 and target DNA sequence and inhibition of DNA binding to p50 by IκBα (an ankyrin repeat protein that can weakly bind to the DNA binding domain). One wheat-germ extract cell-free reaction was used for the expression of the HaloTag-p50 fusion protein, while another reaction was used to co-express HaloTag-p50 and IκBα with T7 epitope tag.
Capture of HT-p50 (Figure 3; slide 1) and HT-p50+ IκBα (Figure 3; slide 1 and 2) complex on a HaloLink slide was confirmed by probing with anti HaloTag antibody and anti T7 epitope antibody respectively followed by AlexaFluor 647 labeled secondary antibody. A dilution series of HaloTag-GST at known concentrations was added to the slide and also probed with anti HaloTag antibody (Figure 3, Slide 1).
A HaloTag-GST titration curve provides an estimate of expression level of HaloTag fusion and is used to normalize for day-to-day variations.
On a separate slide, a p50 consensus DNA sequence labeled with AlexaFluor 647 was used to probe p50 and p50-IκBα dimer. The intensity of DNA-AlexaFluor 647 was threefold lower on p50-IκBα dimer compared to p50 protein alone (Figure 3, Slide 3) validating the result that IkBa binding to p50 DNA binding domain reduces the binding efficiency of consensus DNA sequence. This experiment demonstrates the versatility of HaloLink to not only perform protein-DNA interaction experiments but also extract qualitative information about the binding affinities.

Figure 3. Fluorescence scan showing capture of HaloTag-p50 and its specific binding with its consensus DNA sequence
Conclusion
The HaloLink Protein Array System simplifies custom array creation and offers flexibility and multiplexing capability to researchers looking for an alternative to preprinted protein arrays and their conventional assays such as protein pull-downs and immunoprecipitations. Apart from protein-interaction analysis, HaloLink is useful for researchers looking to do auto-antibody profiling or screening for clinical studies.
Nidhi Nath, Ph.D. ([email protected]), Robin Hurst, and Brad Hook, Ph.D., are senior scientists at Promega.



