March 15, 2012 (Vol. 32, No. 6)
Timothy Galitski, Ph.D. Head of Science and Technology EMD Millipore Institute for Systems Biology
Researchers Increasingly Rely on Holistic Studies to Obtain Greater Insights on Health and Disease
For more than a decade, systems biology has grown rapidly, mainly in academic labs. It is now entering a new phase of commercial opportunity, spreading the practice of systems biology from lab to lab and to the clinic. The biggest challenge facing scientists today is navigating biology’s incredible complexity and applying these insights to clinical medicine.
Traditionally, science has taken a reductionist approach, dissecting biological systems into their constituent parts and studying them in isolation. Entire scientific careers have been devoted to studying only one gene or protein in order to understand its function. Although scientists have made progress using this method, this reductionist approach limits biological insights into the human body. As a result, efforts to treat many complex diseases have also faced limited success.
Reductionism, by its nature, cannot comprehend the complexity of biological systems, the properties of which cannot be explained or predicted by studying their individual components. Scientists now understand that the individual components of biological systems such as molecular pathways never work alone—they operate in highly structured and integrated biological networks.
Health and disease are properties of the changing dynamics of these networks. Thus, to gain a true understanding of health and disease will require a more holistic approach, including the integrated analysis at broadly disparate levels, from molecular to organismal, from genetic to environmental. When scientists unravel this complexity, decipher these dynamic network interactions, and develop predictive models, then they begin to understand life. This ambitious goal demands a systems biology approach.
Today, the biomedical landscape is slowly shifting, moving away from reactive, trial-and-error-based medicine toward what is known as P4 medicine—predictive, preventive, personalized, and participatory approaches to maintaining and restoring human wellness. Molecular testing is already being used by clinicians, particularly in the area of oncology, to help determine which patients will most likely benefit from new treatments and which might experience severe side effects.
Indeed, approximately 10% of all marketed drugs require or recommend genetic testing for optimal treatment and 50% of all clinical trials involve collecting DNA to assist in biomarker development. This shift in the medical landscape is predicated on the capabilities arising from systems biology approaches to biomedicine.
Technological advancements are allowing researchers to take a systems-based approach to understanding health and disease. Whole-genome sequencing technologies, which made the Human Genome Project possible and launched the field of systems biology, hold promise for the assessment of health risk for every individual patient. To monitor health status over time, clinicians will have to supplement genomic information with measures of RNA and protein levels, protein modifications, and other molecular measurements—collectively known as biomarkers—over time.
These measurements require the analysis of many hundreds to thousands of analytes at one time, a process called multiplexing. Together with sophisticated analytical methods and computer modeling, multiplexing facilitates a much broader search for meaningful interactions between the components of biological systems, and enables the analysis of the changing dynamics among these networks between healthy and disease states.
As biomedical research evolves to address biological systems as molecular pathways within complex networks, the tools for analysis must also evolve to allow for this complexity to be revealed, measured, and interpreted. Systems biology is driving the development of handheld or small benchtop tools and platforms that can easily be deployed in clinics or small laboratories to enable systems-level analyses of molecular networks and cells.
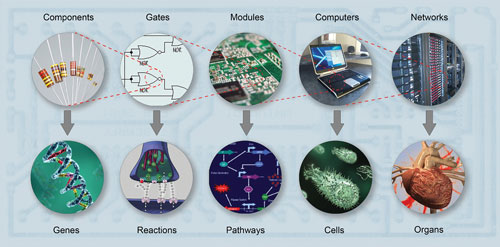
Many systems biologists view aspects of cell biology and electronics as comparable, but with molecules, ions, proteins, and DNA replacing electrons and transistors. Graphic shows correspondence between electronic and biological systems and demonstrates similarities between the components at different levels of complexity. [Equinox Graphics/Photo Researchers]
Many, Faster, Smaller, Cheaper
Multiplexing technologies are the technical core of systems biology. At the genetic level, nucleic acid-based multiplexing technologies, including microarray analysis, multiplex quantitative polymerase chain reaction (qPCR), and multiplex DNA sequencing, are becoming standard tools in laboratories as they allow scientists to analyze the structure and expression of many genes in a single experiment quickly and efficiently. They are frequently used to assist in the identification of disease susceptibility genes and disease-specific gene-expression profiles.
At the epigenetic level—a second layer of hereditary information that causes changes in gene-expression patterns resulting from environmental factors that chemically modify DNA and chromatin—new tools and technologies are available that allow scientists to analyze DNA-methylation patterns, chromatin modifications, and noncoding RNA transcripts. Dysregulated epigenetic modifications are believed to play important roles in the onset and progression of many human diseases, and it has been suggested that these changes may give rise to disease much more frequently than genetic variants.
Although our understanding of epigenetics is at an early stage, the rapid development of these new technologies to define genome-wide epigenetic variations in humans may soon result in effective new epigenetic therapies and diagnostic tests for a wide range of human diseases.
The flow of biological information cuts across networks of nucleic acids, proteins, and metabolites. Thus, disease-specific biomarkers and therapeutic targets are being found in different molecular classes. The fast-growing field of proteomics research requires new tools and technologies that rapidly obtain and analyze systems-level datasets connecting biomarkers and pathways to biological states. This data is impractical to obtain using traditional detection methods, including enzyme-linked immunosorbent assay (ELISA) or Western blotting, because they can only detect up to a few analytes at a time, are semi-quantitative, and offer limited throughput.
Both mass spectrometers and microarrays are being used to measure multiple biomarkers simultaneously, to identify protein-protein interactions and the targets of small molecule compounds in drug development, and to detect the presence or amount of specific biomarkers in human tissue samples such as blood.
This trend of applications development from capital-intensive and somewhat esoteric technologies to more broadly accessible technology will accelerate in coming years.
Multiple scientific developments are working in synergy to advance systems biology. First, new tools and technologies are enabling analysis of the changing dynamics within complex networks that define healthy and disease states. Second, storing and distributing enormous amounts of data has become easier. Last but not least, an expanded ecosystem of scientists are breaking down research silos and working together across disciplines.
Timothy Galitski, Ph.D. ([email protected]), is head of science and technology, EMD Millipore and affiliate professor, Institute for Systems Biology.