January 15, 2015 (Vol. 35, No. 2)
Productivity isn’t everything—expression systems must also deliver quality and robustness.
Mammalian cell-based expression shows an inverse relation between cell proliferation and recombinant protein production. According to Pierre-Alaiin Girod, Ph.D., CSO at Selexis, it is possible to redirect cellular resources from biomass accumulation to protein synthesis by altering the physicochemical environment.
Folding and assembly, however, are highly protein-specific, notes Dr. Girod. Increasing volumetric productivity therefore requires engineering host cells to optimize those functions.
Cells have a fixed amount of critical energy metabolites and ribosomes, resources that are shared between the cellular metabolic pathways. In nondividing cells, all metabolite pools are ascribed to a specialized function, whereas in actively dividing CHO cells, fewer resources are available for producing the transgene.
This trade-off can be controlled by altering environmental conditions—supplying energy-supplying nutrients, such as glutamine and glucose, or using inhibitors, such as butyrate. Recently, investigators have shown that a cell cycle inhibitor can block cell division, thereby redirecting resources toward product synthesis. “Slowing or blocking division also gives more time to the pathways to finish the job as exemplified by N-glycosylation,” adds Dr. Girod.
CHO cells’ ability to divide actively resembles that of tumor cells, and this is why they have become the workhorses of monoclonal antibody production. Transcriptomic analysis shows that out of the approximately 17,000 genes contained in the CHO genome, only 5,700 are actively transcribed. Consequently, many genes have been lost compared with the original hamster genome. “A high number of genes are silenced,” Dr. Girod explains.
Notably, specific steps are limiting. Co-expression of limiting enzymes enabled de-bottlenecking of those pathways.
“Our approach involves engineering of CHO cells by co-expressing multiple enzymes or chaperone proteins that affect specific pathways to create premade libraries. The transgene is then expressed in those libraries,” Dr. Girod says. “Subsequently, clones are isolated from the libraries exhibiting the best productivity and product-quality attributes.”
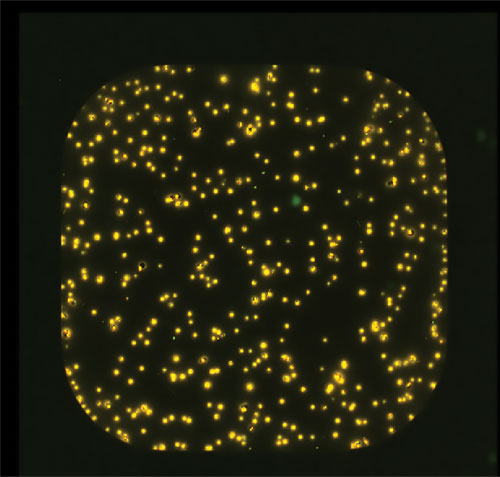
Distribution of mAb secretion within a clonal population at the end of production run. The amount of mAb secreted by each individual cell within a population is quantified by the cell secretion assay. The mAb-sandwich immunocomplexes are stained yellow, and live cells are stained green. [Selexis]
Expressing Human-Like Glycans
As a eukaryotic expression system that is more complex than yeast, insect cell culture has received attention for its ability to fold mammalian proteins properly, thus aiding in their solubility. Insect cells also conduct more complex post-translational modifications than yeast or bacteria. The disadvantages of insect systems relative to prokaryotes include long culture time and higher costs.
Donald Jarvis, Ph.D., president of GlycoBac, believes that insect cells may be improved through strategic knockdowns and gene introductions. GlycoBac, a University of Wyoming incubator company, commercializes technology from the Jarvis laboratory. Specifically, GlycoBac uses the technology to create genetically engineered insect cells for manufacturing vaccines, diagnostics, and therapeutics.
Dr. Jarvis bristles at the term “niche” to describe baculovirus work. “I’m not sure what [the term] means,” he tells GEN. “The baculovirus/insect cell system is now used to produce three FDA-licensed products—two vaccines and an immunomodulator—in the United States and is widely considered to be a good tool for the production of G protein-coupled receptors and other membrane proteins for structural biology.”
Dr. Jarvis’s efforts focus on creating new baculovirus-insect cell systems for improved recombinant glycoprotein production. Native insect cells have simpler glycan processing pathways than higher eukaryotes such as mammalian cells. Dr. Jarvis has modified those pathways by engineering the vector and/or host to produce systems that can be used to manufacture products with humanized or more homogeneous glycan structures.
“We’ve knocked in higher eukaryotic genes and knocked out endogenous functions,” notes Dr. Jarvis. “[These modifications allow us] to obtain human-type glycosylation and eliminate immunogenic sugar epitopes.” Some of the higher eukaryotic genes that have been knocked in are glycosyltransferase genes, which code for enzymes and transporters of sialic acid metabolism. Endogenous genes that have been knocked out include genes that code for immunogenic N-glycans.
Improving Solubility
Lucigen has developed a panel of 24 solubility- and expression-enhancing fusion partners to test multiple tags within a single promoter, vector, and host system. Additionally, the company uses a novel yellow fluorescent protein that enhances solubility and expression while providing visual reports on quantities of soluble protein.
According to Lucigen’s CSO, David Mead, Ph.D., the system allows rapid, high-throughput screening of multiple factors known to improve solubility and expression in the context of an enzyme-free cloning platform.
The fusion partners add to one end of the protein during expression, and are later cleaved using a specific protease site that leaves only the protein of interest.
Although the technique was used here to express a Parkinson’s disease biomarker, it is also applicable to biomanufacturing. “It allows you to screen multiple protein expression tags that have the potential to enhance protein expression and/or solubility,” says Dr. Mead. “Once researchers identify a tag that works, they have the potential to scale up production for biomanufacturing in an Escherichia coli-based system.”
Many proteins are difficult to express in a soluble (that is, functional) format. The Lucigen approach improves the likelihood that such proteins will be expressed in soluble form.
The company plans to release the panel as a kit, the Expresso® Solubility and Protein Expression System, early in 2015. The first-generation kit will use seven tags or enhancers; additional tags will become available through a service-based program where customers work with Lucigen to find a combination that improves expression and solubility levels of their chosen protein.
Expresso enables researchers to create cloning constructs rapidly by mixing amplified DNA with Lucigen vectors and cells engineered for in vivo recombination. “This eliminates purification, restriction, and ligation steps, with greater than 90% cloning efficiency,” Dr. Mead explains. The advantage is that it allows users to create multiple protein expression constructs from a single amplicon in parallel reactions.
Yield versus Quality?
Alpha interferon was the only approved therapy for hepatitis C, but half of patients failed to respond to standard treatment with IFN-α-2b 3 MU, and up to 80% of nonresponders relapsed within six months of treatment cessation. Poor efficacy led to development of “consensus interferon,” or IFN-con.
“IFN-con is a wholly synthetic but highly effective interferon that uses the most frequently observed amino acid in each position from several IFN-α nonallelic subtypes,” explains Ji-won Choi, Ph.D., director of scientific evaluation at PolyTherics. “Hence the name consensus interferon.”
Of the several IFN-con variants synthesized, IFN-con1 is the only variant successfully developed into a product, currently marketed as Infergen® (Amgen). Alpha interferon is currently marketed as PEGylated molecules.
Standard manufacturing methods for IFN-con employ E. coli expression, which is less expensive than mammalian cell culture. PolyTherics also manufactures in E. coli, but to produce soluble protein uses a genetically engineered bacterial strain that contains disruption of the trxB and gor genes, to facilitate formation of two disulfide bonds during cytoplasmic expression. The disulfides help to maintain the protein’s tertiary structure and stability and are essential for bioactivity.
PolyTherics’ improvement consists of no refolding step. “Standard methods produce the proteins in insoluble forms known as inclusion bodies. This process requires a carefully controlled refolding step that aims to regain the protein’s native structure and to pair the disulfide bonds to produce correct folding,” Dr. Choi explains. But refolding operations carry the risk of misfolded protein, a factor that compromises activity, increases aggregation, and raises the risk of immunogenicity.
PolyTherics’ decision to express soluble interferon eliminates the folding steps but is not completely free of drawbacks. As long as inclusion bodies are produced in massive quantities, losing protein during the refolding process is not a terrible loss. “Soluble expression is much more controlled, so you can’t expect folded proteins to be produced to the levels of inclusion bodies, but it’s fully active upon expression and manufacturing is simpler,” Dr. Choi asserts.
PolyTherics uses SUMO (small ubiquitin-like modifier) proteins as fusion partners. SUMO proteins, which covalently attach to other proteins to modify their functions, constitute a unique set of fusion partners. The set is characterized by a chaperone-like property that helps with the correct folding of IFN-con, which occurs prior to the release of a soluble protein complex.
“This helps the native IFN-con to maintain its stability after the removal of the fusion element,” notes Dr. Choi, who asserts that subsequent cleavage of SUMO is simple, specific, and efficient. “Cleaved protein is easy to recover, with the end result being a far simpler process.”

The study of gene expression is closely linked to our understanding of proteins and protein expression. The design of DNA sequences for optimal protein expression is both an art and a science. [Sciencephoto/Shutterstock]
The Meaning of Robustness
We often hear “robust” used to describe processes. What does this mean? According to Hugo de Wit, CEO of Cellca, it means avoiding unnecessary experiments when moving from process development to production: “In the past, new products were viewed as new processes, which caused [users] to repeat experiments on the assumption that everything would be different.”
This view reflects the fact that protein expression methodologies began life as laboratory exercises that were scientifically sound but not easily ported to manufacturing.
“If you walk around a lab, you see highly skilled people behaving as scientists, but in manufacturing you see fewer personnel, more automation,” de Wit says. “Simpler is better, which is not often true in a lab. The requirements for success are different in a lab than in production.”
Cellca’s goal is to leapfrog process development, avoiding it through development-stage processes that scale perfectly regardless of the culture environment. Such super-platform approaches will become necessary, de Wit insists, as personalized medicine becomes routine and the blockbuster model falls by the wayside.
In particular, biopharmaceutical developers will be forced to respond to a combination of greater regulatory scrutiny (particularly with regard to safety), the requirement for companion diagnostics, uncertainty in reimbursement, and anticipated lower revenues per product. Developers will also try to exploit lower development costs, diverting resources to new drugs instead of reinventing the development wheel.
“It’s the same as the automobile industry, which introduces new models based on successful platforms,” de Wit notes.
The Cellca platform is based on the CHO DG-44 expression system, unit operations, and media/feed that work harmoniously. Most of the company’s platform processes achieve 3 g/L titers in a typical 12- to 14-day fed-batch process with little or no tweaking, regardless of bioreactor size or type. “It’s less about productivity than predictability, though,” says de Wit. “The point is to de-risk the protein manufacturing process.”



