October 1, 2009 (Vol. 29, No. 17)
K. John John Morrow Jr. Ph.D. President Newport Biotech
Fast-Breaking Developments Drive Need for Constant Introduction of Novel Technologies
Novel fluorescent tags, new insight into protein crystallization, and alternative expression systems were among the presentations that grabbed participants’ attention at CHI’s “Bioprocessing Summit” held recently in Cambridge, MA.
Green fluorescent protein (GFP) reigns supreme as a molecular detection method. A protein isolated from marine organisms, GFP glows green under wavelengths in the ultraviolet range. When engineered into the sequence of target proteins, it can be used to track cellular events. Geoffrey Waldo, Ph.D., of the bioscience division at Los Alamos National Laboratory, has guided useful modifications of this important technology, working through some of the more vexing issues.
According to Dr. Waldo, “GFP technology is limited by the fact that existing fluorescent protein tags can perturb protein solubility or may not work in living cells.” One way to get around this is to “split” the bulky fluorescent proteins into smaller pieces that might be less bothersome as tags, but existing split GFPs are poorly folded and interfere with protein behavior. This makes them risky to use for monitoring protein interactions and tagging proteins. To overcome these drawbacks, Dr. Waldo and his colleagues have engineered soluble, cell-associating GFP fragments that perform as exemplary tags.
“We evolved superfolder GFP 1-10 by DNA shuffling to improve its solubility and increase its complementation with sulfite reductase GFP 11,” Dr. Waldo stated. The group’s strongest candidate protein had an 80-fold improvement over the starting material. The system is simple to use and works in living cells or in the test tube. Further, the tags do not change the behavior of their target proteins.
Dr. Waldo and his team have used this technology to establish an in situ assay for quantitatively monitoring the aggregation of Tau, the protein associated with neurodegenerative changes in Alzheimer’s patients. This provides a tool for understanding the molecular basis of a number of human neurological diseases.
The Los Alamos team’s split GFP system reportedly lets researchers screen through millions of cells in a few hours to find useful proteins for targets for drug design. A notable achievement from the researchers is a series of split fluorescent proteins, tiny pieces of which are ideal for screening protocols. “The problem with the conventional split GFP protein is that it is plastered with lots of intellectual property protection,” said Dr. Waldo. “We have isolated and engineered an alternative fluorescent protein panel from a sessile coral that sports purple spines.”
With this newly developed scaffolding, an additional tagging option will soon be available to investigators that is free of the intellectual property thicket. “This makes our new split fluorescent protein much more affordable,” stated Dr. Waldo.
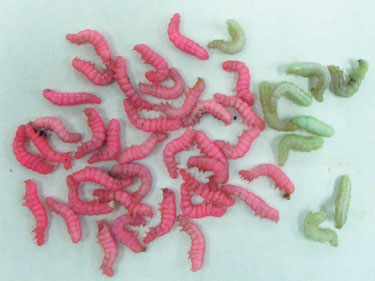
The PERLXpress system from Chesapeake PERL combines well-established recombinant baculovirus technology with mass oral infection of synchronous Trichoplusia ni larvae.
Protein Crystallization
The ocean depths offer unique opportunities for uncovering proteins that have evolved under exotic conditions, according to Joseph Ng, Ph.D., associate professor, University of Alabama, Huntsville. Dr. Ng and his coworkers confront the challenges of developing three-dimensional descriptions of protein molecules through the use of x-ray crystallography.
Current technology takes advantage of structural genomics, in which a sequenced genome is screened for coding regions that can be analyzed through a bioinformatics program. Candidate sequences of interest are then optimized with the goal of expressing proteins and building crystals that can be subjected to analysis.
In a natural state it would be selectively disadvantageous for an organism’s protein to crystallize spontaneously. For this reason, most proteins are resistant to crystallization under normal physiological conditions. Yet, knowledge of a molecule’s 3-D structure is essential for a rigorous understanding of its structure and function, and for the design of drugs that would influence its behavior in a therapeutic situation.
Dr. Ng’s program involves identification, cloning, expression, purification, crystallization, and crystallography of promising molecules. “Success depends on the solubility and stability of the protein and determining the optimal nucleation and crystal growth parameters. Problems may arise if the protein solution is heterogeneous, if the molecules lack a compact arrangement and move extensively in solution, or if the molecules undergo nonspecific aggregation. We have a number of tricks in our bag for overcoming these roadblocks.”
The group has access to a number of hyperthermophilic bacterial proteins, obtained from deep-sea explorations, and these are yielding useful information on the adaptations of proteins to harsh environments. The Huntsville team has found that many of these proteins have a high propensity to crystallize.
Not all heat-resilient proteins are good candidates for crystallization, but for those that are, their structures reveal common molecular features. They possess unique architectural and molecular “hot spots” that endow them with stability and the ability to form crystal lattices. Using high-throughput gene-synthesis and protein-engineering methods developed in Dr. Ng’s lab, proteins that do not crystallize are coupled to hyperthermophilic protein fragments as crystallization vehicles.
Protein crystals suitable for x-ray crystallography can now be obtained by this method where they cannot be acquired otherwise. The subsequent processing of the molecules exploits capillary tubes into which the protein solutions are introduced. The tubes are sealed with nail polish, and counter diffusion takes place within the capillary. The crystals are then subjected to analysis in situ. Many proteins have been analyzed including lysozyme, Factor XIII, Con A, insulin, and glucose isomerase.
“Over 50 percent of targeted proteins in structural genomics programs cannot be crystallized using conventional methods,” Dr. Ng pointed out. “So this approach opens the door to a whole realm that was previously inaccessible.”
Production of Functional Proteins
Elena Kovaleva, Ph.D., head of R&D at Chesapeake PERL, discussed her company’s program of large-scale production of proteins by various lepidopterin species including the cabbage looper. The process involves growing a large number of larvae, infecting them with baculovirus, followed by processing and protein purification.
Cultivation is on a massive scale, generating greater than 10 kg of biomass per hour to a capacity of 200 kg per week for the facility. The company has years of experience with the process, so stability and reproducibility can be assured, Dr. Kovaleva said. By the same token, the baculovirus expression vector system is well-established with custom made vectors based on PERXpress protein production in whole larvae. The work is done with the preoccluded form of the virus, which is infectious when ingested but noninfectious when initiated into cultures in vitro.
Dr. Kovaleva asserted that the heterogeneity of cell types present in the larva allows for a higher fidelity of protein production than that realized with homogeneous cell cultures. Larvae can participate in propeptide and c-terminal cleavage, dimerization, tetramerization, and association with ancillary molecules. Examples of proteins successfully expressed in the PERL platform include fluorescent proteins, viral antigens, virus–like particles, and a range of different enzymes.
A colorful example of the the PERL expression technology is the fluorescent protein DsRed, cloned from the coral Discosoma. Highly stable and bright red, the protein serves as an excellent marker for tracking protein behavior. In order to produce kilogram quantities of protein, infected larvae from the second passage are stored away for later preparation for the next round, thereby overcoming one of the main impediments to large-scale protein production in insect bioreactors.
Baculovirus makes a convenient system for vaccine development, especially given that subunit vaccines are composed of antigen subunits rather than a live virus.
“We are able to continually improve the process, based on host characterization and our broad base of technical experience,” said Dr. Kovaleva. “This places us in a strong position for reaching unlimited production of identical proteins.”
Producing Recombinant Proteins
“In an alternative approach to the use of baculovirus, we have used Spodoptera frugiperda insect cell lines stably transformed to express secreted alkaline phosphatase,” said Satya Prakash, Ph.D., director of the biomedical technology and cell therapy research laboratory at McGill University.
Dr. Prakash said his system offers a number of important features. Recombinant proteins generated in insect cells are glycosylated and biologically active, and large-scale, high cell densities cultures are achievable while maintaining specific productivity. Moreover, stably transformed lines don’t have the problem of protease interferences observed in the lytic system, and the proteins produced are homogenous and consistent from batch to batch. Perfusion and fed-batch modes are easily scalable for insect cell culture, he added.
“We have also generated a potential therapeutic glycoprotein, human Interleukin-7,” he said. This protein is critical for cellular proliferation during B-cell maturation and for T cell and natural killer cell survival, development, and homeostasis. It is an essential growth factor for the immune T cell.
The investigation included a comparison of protein expression using the Baculovirus system with the stably transfected Sf9 insect cell cultures in a conventional stirred-tank bioreactor or the disposable Wave bioreactor system (GE Healthcare), using batch and fed-batch strategies.
The protein synthesized was more homogeneous in the stably transformed cell lines than in the baculovirus-infected cells. Both systems are nonapoptotic and perfect candidates for perfusion and fedbatch systems, he concluded.
Algae excels as a recombinant protein production platform due to its scalability, cost, and containment. In analyzing the economic factor, Dr. Mayfield cited the astronomical yearly costs to patients of a laundry list of protein therapeutics: Avastin $50–100,000; Herceptin $40,000; Erbitux $60,000; and finally Ceredase weighing in at a staggering $500,000. While a number of factors (including corporate marketing decisions) factor into these figures, the estimated financial burden per gram of raw material ranges from $150 for protein produced in mammalian cells down to a minuscule $0.05 per gram for transgenic plants. So it appears that a plant-based protein production platform could only affect consumer prices in a positive fashion.
According to Dr. Mayfield, given selectable markers, transforming plant cells is a relatively easy task, proceeding by homologous recombination. These transformed lines can be built up to commercial scale, at a level of thousands of liters of cell mixtures, in a matter of three to four months. This includes the optimization of properties and the screening and evaluation that accompanies the process. Algal species are GRAS organisms (generally regarded as safe), and although not included in the supermarket gourmet section, they are indeed edible, thus lending themselves to use in vaccine technology.
The organism has another important advantage; it is endowed with chloroplasts that can be easily and conveniently manipulated genetically. Engineering of the chloroplast genome in Chlamydomonas is accomplished through well-described use of promoters and expression elements, and recombinant proteins can be driven to accumulate at high levels. Dr. Mayfield highlighted the use of his team’s technology for codon optimization using GFP markers.
As a proof-of-principle, Dr. Mayfield’s team has engineered strains of Chlamydomonas to produce the bovine serum amyloid protein A3, as well as seven human proteins. This effort included a full-length human monoclonal antibody, which can be shown to accumulate in the chloroplasts.
The use of micro-algae as a biotechnology platform lags behind other organisms, especially bacteria and yeast. This is somewhat surprising given the ability of algae to be grown at large scale in a cost-effective manner. Eukaryotic algae offer tremendous potential for the large-scale and cost-effective production of recombinant proteins.
Fabien Walas, Ph.D., a project leader at ERA Biotech, presented data at the summit concerning the in vivo encapsulation of recombinant proteins in the development of a universal production platform. The technology is based on the zeins or prolaminins, major storage proteins in plants. Specifically, g-zein has the unique property of being soluble in aqueous solution, yet forming aggregates in cells. This process involves a specific, sequential interaction between the various zein species leading to protein body biogenesis. Maize endosperm is especially rich in these large entities.
The Zera® self-assembly peptide was engineered by scientists at ERA Biotech as a fusion protein into which a target of interest was incorporated. Recombinant cells package fusion protein into storage organelles. The Zera fusion peptides interact with the endoplasmic reticulum membrane, inducing the formation of StorPro® organelles, which are insulated from proteolysis. The sequestered proteins are well folded and protected from proteolytic degradation. The platform allows accumulation of protected biomass in a rapid time frame, Dr. Walas added.
The beauty of the system is that the product is protected from the cell, while at the same time the cell is protected from the product and stabilized for long-term storage. This means simpler downstream processing including washing and removal of contaminants.
According to David Wood, Ph.D., a member of the department of chemical and biochemical engineering at Ohio State University, current developments in the bioprocessing industry are driving the need for constant reinvention. These include the completion of the human genome project, the expiration of patents on various biologicals, a massive increase in the yield of recombinant proteins, and a streamlining of the validation process. These changes ensure that there will be a continuing demand on companies and academic investigators to bring forth updated versions of their favorite bioprocessing technologies.
K. John Morrow Jr., Ph.D. ([email protected]), is president of Newport Biotech and a contributing editor for GEN. Web: www.newportbiotech.com.