April 15, 2010 (Vol. 30, No. 8)
James Netterwald, Ph.D.
Better Tools and Methodologies Are Needed to Move the Field Past a Critical Stage
Microarrays have advanced significantly over the years, which is a good thing as genomics would not exist if not for that progress. Protein microarray development, however, has not seen as much success as DNA microarray development, which has been somewhat worrisome for proteomic scientists—and life scientists, in general. In fact, it is likely that the genomics revolution may come to grinding halt if protein microarray development does not catch up soon.
If the focus on protein microarrays at many of this year’s scientific conferences is any indication, academic and industry researchers are aware of the dire need for progress in the field and are endeavoring to deliver the cutting-edge methodologies and tools that will help to avert a crisis.
Yashu Liu, Ph.D., a postdoctoral fellow at the University of Michigan (UM), spoke at “PittCon” about development of a protein microarray to perform biomarker screening in serum from hepatocellular carcinoma (HCC) patients. From this screen, UM researchers identified five biomarker candidates (seroplasmin, Histidine-rich glycoprotein, Complement C3, CD14, and hepatocyte growth factor) that were initially discovered using mass spectrometry and then validated using the protein array.
To create the array, Dr. Liu raised monoclonal antibodies against known serum proteins and bound them on a nitrocellulose-coated glass slide. The screen was then performed by incubating the array with HCC patient serum to screen for potential HCC biomarkers (antigens).
Biotinylated lectins were then used to detect glycan structures on the antigens in the serum, followed by incubation with a fluorescent dye to detect the biotin. “We developed this method so that people could use it with any kind of cancer. If you think there will be a glycan structure change in cancer antigens, then you can apply this process to another type of cancer or another disease,” said Dr. Liu. “Alphafeto protein is not good enough for early detection of HCC so the proteins we found could potentially aid in the detection and diagnosis of this cancer.”
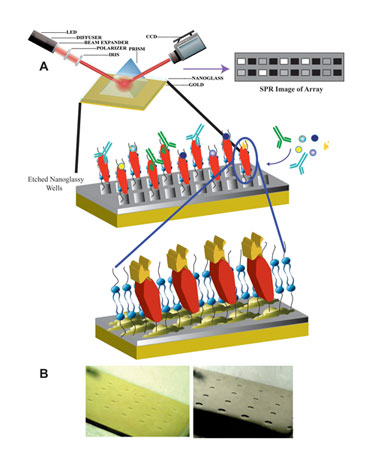
Matthew Linman, a graduate student in Quan Cheng, Ph.D.’s research group at the University of California, Riverside, reviewed currently unpublished data on the initial development of a lipid microarray designed to immobilize transmembrane proteins at “Pittcon”. SPR was used as a functional assay for proteins immobilized on the array. The array, which is based on a calcinated chip, consists of a thin layer of glass atop a layer of gold.
There are several reasons for this design. One reason is that, in order for SPR to work, a gold layer is required. Lipid vesicles, however, do not fuse directly to gold but will fuse to glass. By combining both elements in one array, the group was able to achieve lipid fusion and successful in situ protein function results via SPR.
“We are currently designing the surface, which is called a biological interface, to mobilize epidermal growth factor receptor (EGFR),” said Linman. “The idea behind using this array is to create a surface to test out small molecule inhibitors of EGFR to give patients with head and neck cancer more options than just monoclonal antibody-based therapies such as Erbitux.”
For the last five years, Peter Nilsson, Ph.D., associate professor at KTH Royal Institute of Technology, has been developing various types of protein arrays. Dr. Nilsson’s lab has produced two kinds of protein arrays—antigen arrays for antibody validation, as well as suspension arrays, which are color-coded beads with immobilized antibodies, to perform plasma profiling.
“Our goal is to take all of the antibodies that come out of the Human Protein Atlas project and profile them on serum or plasma cohorts,” said Dr. Nilsson. “We increased the number of antibodies and samples that can be used for simultaneous profiling by direct-labeling assay based on a biotin-streptavidin interaction.”
The main purpose of this array is to discover new biomarkers by comparing plasma and serum samples from various disease cohorts. Dr. Nilsson will present a massive number of data points generated with antibodies profiled on the samples at Select Biosciences’ “Advances in Microarray Technology” to be held in Dublin next month. His presentation will show the “possibility of using a large number of antigens and antibodies to identify biomarkers of disease in a discovery context.”
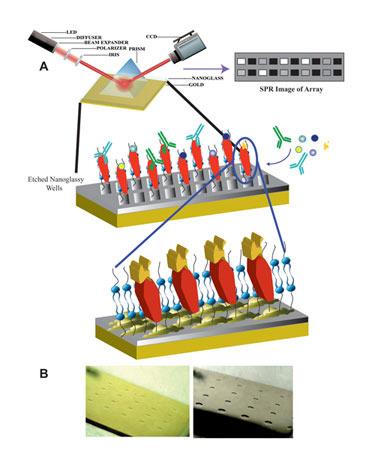
Researchers at University of California, Riverside are working on a lipid microarray designed to immobilize transmembrane proteins. (A) Schematic of membrane protein array with SPRi detection. (B) Optical images of 4 x 5 etched array patterns of squares (left) and circles (right).
Biomarker Measurement
Jim Rusling, Ph.D., professor of chemistry and cell biology at the University of Connecticut, is pursuing the development of arrays for the accurate measurement of cancer biomarker proteins for early cancer detection, surgical applications, and cancer monitoring. At present, researchers in Dr. Rusling’s lab are working on various approaches that can do this in an ultrasensitive fashion.
In one of those methodologies, Dr. Rusling uses nanostructured sensor surfaces as a basis for immunosensor arrays. The nanostructured surfaces could contain carbon nanotubes or tiny gold nanoparticles, and they have chemistry that allows the user to attach antibodies to the cancer biomarkers.
“The antibodies capture the protein of interest from the sample, and then we come in with a labeled secondary antibody that is attached to a multiple enzyme-labeled nanoparticle,” he explained. “We are also interfacing these arrrays with microfluidics for automated use.”
Dr. Rusling has reportedly been able to achieve extremely good sensitivity and detection limits. “We hope that the devices will be used in doctors’ offices and clinics for screening patients.”
New Tools
With regard to new array design tools, PEPperPRINT has developed a new biochip platform with which customized high-density peptide microarrays can be generated at reasonable costs, said Volker Stadler, Ph.D., CEO.
To create these arrays, PEPperPRINT employed xerography—a method in which the company “prints amino acid toners by means of a 20-color printer on conventional glass slides.” Melting these toner particles releases the embedded amino acids for high-density combinatorial peptide synthesis, he noted.
Dr. Stadler will talk about xerography at the Select Biosciences’ meeting. He will cover the basics of this new approach and present case studies that demonstrate the usefulness of peptide microarrays generated. “These case studies cover different applications from epitope mapping and antibody profiling up to the identification of new and unnatural target binders and maybe also some enzymatic assays.”
Larry Gold, Ph.D., CEO and chairman of the board at SomaLogic, will present data on a new class of reagents called SOMAmers at the Select Biosciences meeting. Created using a form of DNA, SOMAmers “are effectively super-monoclonals that allow one to bind to proteins in plasma and serum with nearly absolute specificity,” Dr. Gold reported.
The reagents were developed with the aim of eliminating the problem of high noise that is intrinsic to the use of monoclonal antibodies in array-based detection and were used to build a protein microarray for biomarker discovery.
SomaLogic’s array is essentially the result of learning from the mistakes of past attempts at developing protein arrays. The assay increases the specificity of array-based protein detection by decreasing binding avidity (by performing all binding reactions in solution rather than on printed slides), Dr. Gold added.
“Finally, at the end of the assay, because SOMAmers are made out of DNA, they can be quantified on a DNA chip—in fact we quantify proteins by measuring the SOMAmers that are bound specifically to each protein.”
Dr. Gold’s presentation will focus on the absolute specificity and high precision of these arrays as tools for protein biomarker identification and quantification, as well as show clinical data that demonstrates the array as a potential diagnostic assay for non-small-cell lung cancer.
When it comes to biomarker development, researchers have not even begun to scratch the surface, especially when trying to predict disease. “Using a single biomarker, the statistics of cancer prediction success are not very good. So the advantage of having an array and measuring a number of biomarkers, which could be as little as four or as many as 20, is that the statistics of prediction goes way up. And some people say that, if we are able to do this, cancer prediction may reach 100 percent. That means the reliability of your screening process will be much improved compared to current use of a single biomarker,” Dr. Gold said.
Microarray development is a relatively new science, with much of the information known for DNA arrays and protein arrays trying to play catch-up. The work being done in the field will likely bring protein arrays closer to realizing their full potential.