October 1, 2010 (Vol. 30, No. 17)
Vicki Glaser Writer GEN
Technique Gaining Ground and Leading to Novel Insights on a Genomic Scale
Over the past several years, RNAi screening went through a period of technology development aimed at gaining a better understanding of its advantages and limitations, and there is now an “established set of guidelines and good practices,” says Steven Haney, Ph.D., an associate fellow in Pfizer’s applied quantitative genotherapeutics unit.
Development in the field is now more focused on how RNAi technology can be used to probe fundamental biology, he observes. One example is Pfizer’s use of siRNA image-based screening to identify kinase genes that allow cancer cells to adapt to and survive in a low-oxygen environment. Dr. Haney will be one of the presenters at CHI’s upcoming “RNAi for Functional Screens” conference.
Unlike proliferative cancer cells that grow unchecked in the oxygen-rich periphery of tumors, cells in the interior of a tumor must adapt to an acidic and hypoxic environment. It is in this interior compartment that researchers believe metastatic cells acquire their ability to leave the primary tumor and where some sort of tumor stem cell or tumor-initiating cell is believed to reside, at least in some tumor types.
Under limiting oxygen conditions, cells adapt to the hypoxic environment by undergoing various types of changes, including aberrations in nuclear morphology.
To identify which genes in the kinase family are associated with these changes in nuclear morphology, Dr. Haney’s group designed a high-content screen that incorporates multiple siRNAs per kinase gene and uses 4´,6-diamidino-2-phenylindole (DAPI)—a fluorescent stain that binds to DNA—to visualize changes in the size and shape of nuclei. They chose siRNA screening because it yields information on the direct link between gene expression and cellular adaptability to hypoxic stress.
“RNAi screening is a very valuable method for collecting information about what controls a process,” he says. To generalize their findings, his group is screening multiple different cell lines from tumor types known to leverage their ability to adapt to hypoxic stress to drive their aggressive growth. These include colon cancer, renal cell carcinoma, and melanoma.
A critical bottleneck in cancer research and therapeutics development has been translating the behavior of tumor cells grown in culture to what occurs in vivo, in Dr. Haney’s view. Advances in high-throughput screening strategies aim to “push the biology,” he says, and to develop methods for doing in vitro screening in a setting that most closely models the in vivo environment.
High-content assay technology is creating new opportunities for studying cells using biological assays to measure more subtle changes in cell activity and viability than traditional apoptosis endpoints.
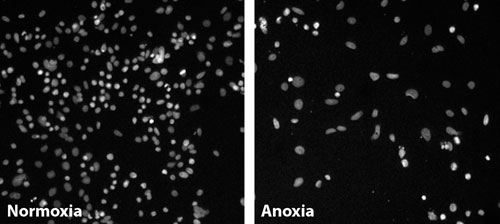
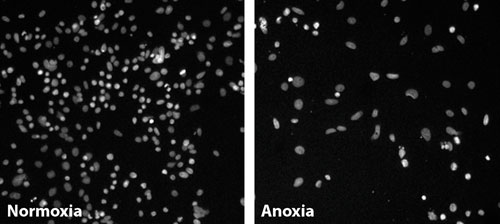
Nuclei of colon cancer cells grown under normoxia and anoxia (>1% oxygen): The difference in nuclei shape and texture between these two conditions formed the basis of a screening endpoint to identify genes that interfere with how cancer cells adapt to the hypoxic environment within a tumor. These differences were quantified through digital imaging and automated analysis. siRNAs that caused statistically significant differences were noted, and genes with several siRNAs causing such changes were identified as hits. [Pfizer]
Genome-Wide Screening
To pursue his interest in understanding the biological basis of genomic instability and DNA damage repair systems, Alex Bishop, D.Phil., assistant professor, cellular and structural biology, at the University of Texas Health Science Center, is using RNAi screening to probe cell survival and response mechanisms following exposure to DNA-damaging agents.
A cell’s DNA-damage repair system coordinates a multifaceted, pleiotropic response that involves multiple pathways affecting, for example, cell-cycle regulation, cell growth, DNA repair, transcription and translation control, and proteasome degradation. The result may be a controlled shut-down of cell function leading to apoptosis, progression down a path of DNA repair and the return to a growth state, cell senescence, or some intermediate response.
Because so many cellular processes are involved, “this is a perfect situation to look at the entire genome,” says Dr. Bishop. And RNAi screening in Drosophila offered a means to do just that. By exposing Drosophila cells to DNA-damaging agents and introducing RNAi reagents to achieve targeted gene silencing across the genome, Dr. Bishop is hoping to identify genes—and the pathways in which they function—that are activated when cells carry out DNA repair.
This work has a direct translational component, as many existing chemotherapeutic drugs used to treat cancer exert their effects by causing DNA damage in dividing cells, impeding their ability to replicate. How these drugs work and how cells develop resistance to them is not well understood, and Dr. Bishop believes that these types of RNAi screening studies will help shed light on the critical mechanisms involved in these processes.
Dr. Bishop has had to overcome several challenges in applying RNAi assay technology for whole-genome analysis in Drosophila. The first is the technology’s intrinsic propensity for off-target effects. About 40% of the data is “real” and readily validated, he notes, whereas the remaining 60% may be junk or at least of questionable utility.
This same issue plagues mammalian RNAi screening. Another stumbling block has involved translating the information obtained in Drosophila to humans, as most of the bioinformatic tools available for genomic analysis were developed for human data and most existing orthology mapping tools were inadequate.
Dr. Bishop has adapted the RNAi assay system he is using to a robotics system to allow for high-throughput screening. To enable better comparison of data between microtiter plates and to help normalize the data and reduce plate-to-plate variation as well as plate location effects, Dr. Bishop’s group loads samples onto the plates in a distributed pattern, interspersing siRNA across the plate and utilizing 10% of the plate’s real estate for controls. This strategy has increased the stringency and sensitivity of the screen.
In addition to mining the screening data to look for individual genes linked to DNA repair and cell survival, another component of Dr. Bishop’s data-validation strategy is to do pathway analysis. The ability to link multiple genes in a pathway to a cellular activity of interest increases one’s confidence in the strength of that association, Dr. Bishop explains.
He is also probing the protein interactome to support data validation and minimize the risk of false-negative results. By combining three databases that cover five species, he is able to compare interactions across species and to configure protein interactions into pathways. This has helped to simplify the interactome, making it a more useful tool for data mining and intuitive gene discovery.
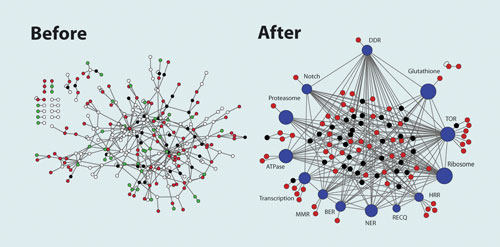
A simpler and more intuitive interactome can be produced by integrating pathway information into a protein interactome analysis of genomic data. [University of Texas Health Science Center]
Thermo Fisher Scientific conducts in-house RNAi-based screens as a service and to assess best practices, evaluating screening-design strategies and hit-stratification methods.
James Goldmeyer, Ph.D., a field application scientist at Thermo Scientific Genomics, a Thermo company, describes a study designed to evaluate different screening strategies, incorporate siRNA reagents that minimize off-target and secondary knockdown effects, avoid the selection of promiscuous molecules, and optimize data interpretation and validation. “I don’t think we will ever completely eliminate the off-target events, but there are ways to mitigate them and follow-up strategies that allow for easier interpretation of the data,” says Dr. Goldmeyer.
In one screen, Thermo scientists utilized a ubiquitin-EGFP assay designed to identify druggable genes required for proteasome function. They selected a proteasome screen because it is well studied and well understood, relevant gene ontology tools are available for performing whole-genome searches, and the assay design is amenable to multiplexing.
The question they wished to examine is the following: is it better to “start out with a broad assay design and then qualify candidate hits with a more specific assay (such as cell death followed by pathway analysis), or can you start out with a specific assay and then drill down even further (for example, a cell-based assay followed by target-specific assays)?” They tested two different assay strategies and two siRNA design approaches.
The assay strategies included a common cell-viability assay and a pathway-specific GFP fusion assay that directly targets proteasome degradation. The results indicated that “the more specific the assay, the more specific the data,” Dr. Goldmeyer says. Both approaches yielded “high-confidence candidate hits”; the screeners’ design choice, he concludes, will depend on factors such as time, reagent and consumable costs, and how many false positives they are willing to accept to minimize the risk of missing a false-negative result.
One common practice is to develop an assay using a pooled reagent for the primary screen followed by deconvolution of the pool and rescreening of candidate hits using individual constructs. The screen described by Dr. Goldmeyer incorporated two genomic siRNA libraries, one of which was chemically modified for enhanced specificity. The screens were run in parallel as gene-specific pools and the individual constructs that made up these pools.
The first approach—screening with pools and then with individual siRNAs—yields more data points per gene, and the number of individual construct hits can be rank-ordered based on increasing confidence. The alternative approach offers higher throughput and generates a smaller number of yes/no data points; multiple positive results indicate higher confidence.
The outcomes for various combinations of reagents yielded a hit list with overlapping high-confidence candidates. Cost/benefit considerations and whether users are more comfortable applying bioinformatics tools to a large dataset or working with less data in which they have greater confidence regarding their validity will drive screening design decisions. The specificity of the assay and how much noise is in the system will also be contributing factors.
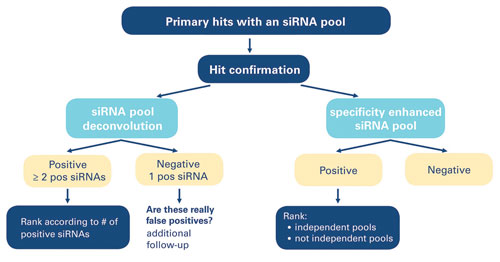
Thermo Scientific Genomics’ scientists rely on a well-defined reagent selection and screening strategy: Available options are to use the parallel-pooled reagents, individual constructs from the two libraries, or combinations of pools and individuals in parallel or sequentially. The strategy one chooses to employ will require differing screening workflows and analysis methodologies, but all will yield high-quality hits, according to the company.
Linking Genes to Pathways
Hyperactivity of the Wnt/beta-catenin signaling pathway has been associated with several forms of cancer. Furthermore, diseases such as osteoporosis and Alzheimer disease have been linked to decreased beta-catenin activity. A research team at Merck & Co. led by Michele Cleary, Ph.D., senior director of automated biotechnology, designed a lentivirus-based short hairpin (sh)RNA screen to identify genes involved in the regulation of Wnt/beta-catenin signaling.
The large-scale screen tested a 14,000-vector shRNA library against 5,000 human genes. The assay was based on a luciferase reporter of beta-catenin driven transcription that was developed in the laboratory of Randy Moon at the University of Washington.
The Merck team’s screening strategy involved pretreating the cells with an inhibitor of a naturally occurring protein complex containing GSK3β (a serine/threonine kinase) that acts to downregulate beta-catenin activity.
“We treated the cells with a dose of the GSK3β inhibitor that was just below the level needed to detect a change in reporter activity. By taking this approach, we were able to screen for genes whose RNAi-mediated loss synergizes with this low dose of GSK3β inhibitor to potently activate the reporter,” explains Dr. Cleary.
She describes two main advantages of using lentivirus delivery of shRNA versus transfection of siRNA: improved delivery efficiency in cell types that are difficult to transfect, and the ability of lentiviruses to integrate into the host genome and provide long-term gene silencing in dividing and nondividing cells. The Merck group developed protocols for preparing thousands of lentivirus constructs in parallel in microtiter plates at a reasonable cost.
From the screen they identified “new modulators of the beta-catenin signaling pathway, most notably dihydrofolate reductase (DHFR).” Follow-up studies showed that DHFR inhibition decreases GSK3β-mediated phosphorylation of beta-catenin, leading to accumulation of beta-catenin in cells.
Dr. Cleary points out that methotrexate, a drug used to treat inflammation, targets DHRF; GSK3β inhibition appears to diminish the inflammatory response; and the actions of methotrexate and the GSK3β inhibitor are synergistic. These observations, combined with evidence that patients with rheumatoid arthritis who are treated with methotrexate have an increased risk of certain cancers, highlight the potential for certain types of anti-inflammatory agents to activate a potent oncogenic pathway.
The key challenges in applying RNAi technology “lie in applying the appropriate resources to the sometimes extensive follow-up needed to understand how a particular gene feeds into the relevant pathways and how this information can help advance a drug discovery program,” says Dr. Cleary.
Particularly if it is not possible to screen in replicates—for example, “when working with large RNAi libraries or expensive assay readouts.” Follow-up screens to confirm hits and ensure on-target silencing are critical for a successful screening campaign, she adds.
The Translational Genomics Research Institute (TGen) is applying high-throughput, high-content RNAi to advance a systems-biology approach aimed at mapping perturbations across the transcriptome to molecular networks and using this information to support iterative testing and refinement of models of gene activity related to molecular and cellular processes.
The goal of this effort, says Spyro Mousses, Ph.D., director of the Center of BioIntelligence of TGen, is to develop “a more functional way of interrogating the human genome and assigning a trait to a gene.” The advent of siRNA libraries offered a means to look across the entire genome and identify groups of genes that converge on a particular phenotypic profile.
Key technology improvements will include continued miniaturization of assays beyond the current 384-well format, which will help further reduce the cost of screening, notes Dr. Mousses. He describes the ability to marry high-content and high-throughput RNAi screening as one of the most significant innovations enabling the simultaneous collection and assessment of multiple response parameters.
Advances in informatics, with the availability of more powerful mapping and knowledge mining tools, are facilitating data interpretation and helping researchers identify multiple genes linked to a phenotype and group them into pathways. This information has a range of applications, including target discovery, pharmacogenomics, probing cellular response to a drug, and biomarker discovery.
Dr. Mousses emphasizes the importance of “understanding the limitations of what inferences you can make from the data,” learning how to minimize off-target effects, artifacts, and false-positive or -negative results, and recognizing the risks of pooling siRNAs. “These limitations do not decrease the value of the technology,” he adds, and there are many strategic approaches for dealing with them.