February 1, 2015 (Vol. 35, No. 3)
Scientists Learned to Move Beyond One-Drug, One-Target Methods of Thinking
As pharma’s R&D productivity crisis continues, more researchers are asking whether there hasn’t been too much reliance on target-based screening methods at the expense of empirical experimental biology.
“Perhaps we were expecting too much from the single-target paradigm,” says Andreas Vogt, Ph.D., an associate professor of computational and systems biology at the University of Pittsburgh.
An article in the July 2011 issue of Nature Reviews Drug Discovery described a renewed interest in phenotypic screening. The goal of this technique is to identify substances such as small molecules, peptides, and RNAI that can alter the phenotype of a cell or an organism in a particular manner.
In the journal article, David Swinney, M.D., CEO and cofounder of the Institute for Rare and Neglected Diseases Drug Discovery, and Jason Anthony, then at Roche in Palo Alto, found that 28 of the 50 first-in-class small-molecule drugs approved by the FDA between 1999 and 2008 had been discovered with phenotypic rather than target assays. The authors asked whether a too “target-centric” approach to drug discovery might be contributing to high attrition rates and lower R&D productivity.
Since the article appeared, more laboratories have been stepping up their use of phenotypic screening, which is being updated through use of surrogate cell studies, robust cell-line development, imaging and analytics technology, and high-throughput target prioritization.
“The idea is to adjust the discovery paradigm,” continues Dr. Vogt. “Combine the benefits of high-throughput approaches with those of more relevant biological approaches.”
Imaging and analytics figure prominently in some new tools for carrying out phenotypic screening. Animated Dynamics (Anidyn), for example, has developed a biodynamic imaging platform that uses holography and laser radar to measure changes in human tissue in real time. The technology, which offers a clear view of how tumor and other cells respond to different drugs and drug dosages, has undergone preclinical testing and will soon be tested in human clinical trials. The company, which licenses the technology from Purdue, expects to receive regulatory approval and to launch commercial sales by 2017.
David Nolte, Ph.D., physics professor at Purdue and president of Anidyn, says the platform will provide a biologically relevant context to lead selection. “The biology happening in petri dishes during lead testing is not the biology that goes on within tissue, notes Dr. Nolte. “There are differences in how cells respond to drugs in a three-dimensional environment, which means the results that occur in Petri dishes may not be the same as the results that occur in the body.”
“We use spectroscopy to measure the time-dependent changes in the hologram,” adds John Turek, the company’s executive vice president and CFO. “It breaks down the changes into different frequencies, and we can tell how a cell’s membranes, mitochondria, nucleus, and even cell division respond to drugs. We measure the frequency of the light fluctuations as a function of time after a drug is applied.”
The platform makes digital holograms of tissues, allowing for visualization, not just at the surface, but throughout the tissue. The technology can also visualize the effects of drugs over time. It will be marketed for use in personalized cancer treatment to help physicians develop the most effective treatments and as a companion diagnostic to optimize patient selection for clinical trials.
In the laboratory, the imaging is compatible with different tissue formats, including tumor spheroids grown in bioreactors or in multiwell plates, as well as tissue biopsies and other organotypic models. It can be used in assays for chemosensitivity and resistance and toxicity screening.
This year, Anidyn will introduce new components and software that will help users integrate its platform with Zeiss and Nikon inverted microscopes. The idea is to allow standard issue microscopes to show changes in a cell’s phenotype in real time.
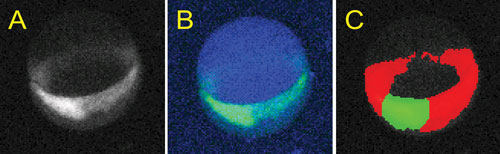
These images, provided by the University of Pittsburgh Drug Discovery Institute, show zebrafish embryos being used in drug discovery: (A) an organogenesis gene expressed in a developing zebrafish embryo at 7 hours post fertilization; (B) a custom-designed image analysis algorithm based on Definiens Cognition Network Technology; and (C) the transgene identified within the embryo (blue), segments shield (green), and germ ring (red) for quantitation.
Affinity Chromatography
Analytical methods such as chromatography are also coming into play. Shantani, a chemical proteomics company based in Mumbai, has developed an affinity chromatography method designed to identify targets from phenotypic screening and to help clarify the mechanism of action.
According to CEO Chaitanya Saxena, Ph.D., Shantani’s products are a subcellular location specific (SCLS) target-capture technology and a unique polymer technology (UPT). These technologies, which identify targets from phenotypic screening hits, differ significantly from current target identification approaches that require substantial amounts of structure-activity-relation (SAR) information on hit molecules, notes Dr. Saxena, who adds that with UPT, hits can be used directly for target identification.
According to Dr. Saxena, Shantani’s approaches to identify targets and trace mechanisms of action are significantly faster, simpler, and more cost-effective than alternatives. He claims that the company’s UPT, for instance, can generate target identification information in as little as three weeks.
Shantani is using these methods in its own drug development efforts, which already have identified a new target for a metabolic disorder. The firm expects to shortly start IND-enabling studies on optimized lead molecules for a type-2 diabetes target. The company is also collaborating with a top 10 pharmaceutical company on an oncology drug—a non-kinase, non-VEGF, non-vascular disrupting agent for anti-angiogenesis therapy, points out Dr. Saxena.
He admits that the company’s SCLS methods are limited to cellular models. They have not been used in animal screens, and they have not been tested in screens that involve neuronal cultures.
Targeting Alzheimer’s Disease
Phenotypic drug development is also proving important for an area where many drug discovery and development projects have failed: Alzheimer’s therapies. Ultimately, the field must be driven to target-based screening, says Tae-Wan Kim, Ph.D., associate professor of pathology and cell biology at Columbia University Medical Center. But, to date, not much high-throughput screening has been done.
“The data is there for cancer, but with Alzheimer’s, we’re not there yet,” cautions Dr. Kim, citing a clinical trial success rate for Alzheimer’s drugs of 0.4%.
Phenotypic methods will be necessary to better understand Alzheimer’s and other complex disorders, he continues, adding that his teams are using phenotypic, high-throughput screening platforms targeting cellular pathways to Alzheimer’s, including beta secretase (BACE 1) inhibitors, Tau protein modulators, and apolipoprotein E (ApoE) enhancers in primary and stem cell-derived neurons and glial cells. The lab is currently working on 384-well throughput systems, measuring protein levels and dendrite mis-sorting in a variety of cells.
Dr. Kim and colleagues are also looking at identifying identify existing drugs that might be repositioned as Alzheimer’s treatments. For APoE enhancers, the lab has screened over 15,000 compounds with a hit rate of 1.8%.
Cell Assays
Other research is tackling another major problem with phenotypic screening: developing relevant assays in human tumor cells and other cell cultures. One major problem is actually growing the cultures, which involves ensuring that a sufficient number of cells are available for assay, since cells often lose their ability to reproduce.
Propagenix, a company set up last April, is developing the conditional reprogramming (CR) cell technology, which allows epithelial and tumor cell cultures to grow in a stem-cell-like state, and to return to their normal state once removed from the culture media. The technique involves adding irradiated fibroblast feeder cells and the rho kinase (ROCK) inhibitor, Y-27632.
CR was developed by virologist Richard Schlegel, M.D., Ph.D., chair of the pathology department at Georgetown University Medical Center’s Lombardi Cancer Center, and one of the inventors of the human papilloma virus (HPV) vaccine. In 2011, Dr. Schlegel was developing a new tissue culture to study the HPV virus, explains Brian Pollok, Propagenix’s CEO.
The company’s new lab has been working in parallel with Schlegel’s labs for the past six months. Research on the platform has also been underway at the NIH and the National Institute of Allergy and Infectious Diseases, using breast and lung cancer tumor cells.
Serendipity played a part in developing the technology, Pollok says. While working with epithelial cells, Dr. Schlegel noticed that they grew in colonies, characterized them, and saw that they took on some of the properties of stem cells without the kinetic effects often found in some stem cells. Researchers have reported using the platform and being able to grow cultures from as few as four cells.
At this point, emphasizes Pollok, the most exciting application is going on with airway epithelial cells: “You expand out from biopsies, and when you take the cells out of the CR media, they redifferentiate, become polarized and even have beating cilia, showing that you can take normal cells, grow them, and put back redifferentiated cells that go back to their initial state.” The platform only works with epithelial cells, he says.
The CR platform also ensures verifiable biopsies for cultures. “You tend to get both tumor and normal cells in biopsies, and both will proliferate, until normal cell growth can outpace that of cancer cells,” Pollok explains. “Then you need to separate out normal from tumor cells, or detect them based on biomarkers. We are using flow cytometry to do this, determine the functional differences and the pathway activated in tumors, and then use reporter genes.”
The CR platform is faster and less expensive to use than traditional methods, according to Pollok, and Propagenix is working not only with airway epithelial but pancreatic islet cells. Applications would include culturing for phenotypic screening, but also personalized tumor chemotyping and, potentially, cell therapy.
“In theory, the platform could allow one to take patient-delivered material, grow it out, genetically engineer it, then use gene editing to correct problems, and transplant the corrected cells back in,” suggests Pollok. “Partners are looking into to how they might do just that.” At present, the company seeks discovery research collaborations with pharma and biopharma companies.
Another basic challenge for phenotypic screening is developing cell systems relevant to humans in surrogate creatures. Dr. Vogt and his team at the University of Pittsburgh have been working with zebrafish phenotypic screening to develop a quantitative systems pharmacology approach that could enable the discovery of new treatments for acute kidney injury.
The problem with these injuries, he says, is that they result in the formation of scar tissue, and effects are often advanced by the time a patient seeks treatment. Dr. Vogt’s team found the zebrafish’s kidney regeneration offered a good model.
They developed a phenotypic platform for automated imaging of transgenic zebrafish embryos. Taking advantage of Lim1 homeobox protein Lhx2, the system uses enhanced green fluorescence protein EGFP-tagged marker to trace the impact of drugs. It uses cognitive network technology to allow visualization of the marker, which is expressed during kidney development but also after injury, as tissue is regenerated.
So far, their efforts have found a series of molecules that could aid the repair process and reduce the formation of scar tissue.
Research has given them an explanation of why and when, but the fundamental question remains. “What we don’t know is how they do this,” says Dr. Vogt. “Something is needed at the back end to let us test how compounds affect activity, since we’re starting with a system that is hard to focus on, or isn’t monogenic, where we don’t know the target.”
Possible options include growth factor signals, genetic manipulation, and hypothesis based on models to figure out pathways. There could be a role for chemical proteomics, as well as mass-spec-based methods, both of which are going on at a number of universities. “In the end,” concludes Dr. Vogt, “you want to find the target, or at least you want to know more about the mechanism.”

Conditional reprogramming technology, developed in the Georgetown University laboratory of virologist Richard Schlegel, M.D., Ph.D., allows epithelial and tumor cell cultures to grow in a stem-cell-like state, and to return to their normal state once removed from culture. The technology is now being developed by Propagenix.
High-Content Analysis of 3D Microtissues
3D cell culture models are widely accepted as being more physiologically relevant than 2D methods and a promising tool to improve the drug de-risking processes. The current standard to gain high-content information on a cellular level within tissues is histological analysis, but this impedes 3D model use in early stages of drug development.
High-content analysis (HCA) is HTS-compatible and allows similar quantity and quality datasets to be gained, thus enabling the use of 3D models early on. However, HCA cell visualization in 3D models is challenging because light scattering and absorption prevent imaging of regions deeper in the tissue.
PerkinElmer designed the Opera Phenix™ high-content screening system to study complex cellular models such as 3D microtissues. Its Synchrony optics can provide excellent confocality and enable simultaneous acquisition of up to four channels with minimal crosstalk. The confocal microscope spinning disk has a special design with an increased pinhole-to-pinhole distance for reduced spatial crosstalk from out-of-focus planes.
“This results in clearer images of 3D microtissues,” said Karin Boettcher, associate product manager, cellular imaging and analysis.
To demonstrate the Opera Phenix system’s use in drug discovery and preclinical safety assessment, the company developed a high-content imaging assay to quantify the efflux transporter activity in 3D human liver microtissues provided by InSphero. A functional impairment of hepatobiliary transporters such as bile salt export pump (BSEP) and multidrug resistance-associated protein 2 (MRP2) are strongly associated with an increased risk of liver injury.
“Currently, artificial models, such as BSEP expressing membrane vesicles, are usually used for studying efflux transporter function,” continued Boettcher “But they lack the functional complexity of the natural 3D liver environment that the the Opera Phenix system is able to uncover.”
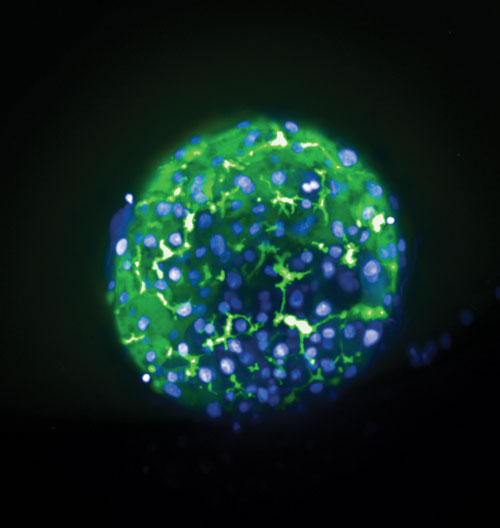
3D human liver microtissues from Insphero were labeled with Hoechst (blue, nuclei) and CMFDA (green) and imaged on the Opera Phenix HCS system (20xW, Max Projection of 60 µm). CMFDA is exported by hepatocytes into bile canaliculi and labels the bile canaliculi network. [PerkinElmer]



