March 1, 2013 (Vol. 33, No. 5)
The development of robust, cost-effective molecular tools for identifying genetic variants and mutations that underlie human diseases requires innovative approaches.
Medical diagnostics for the detection of such variants or mutations facilitate attempts to individualize patient treatment and prognosis. Most currently available molecular assays are limited in their detection sensitivity for low-abundance genetic variants of disease.
A number of researchers engaged in the field of nucleic acid sample preparations are scheduled to present their techniques and methodologies to enrich regions of interest in the human genome at CHI’s upcoming “Sample Prep and Target Enrichment in Molecular Diagnostics” conference.
The analysis of proteins and biomarkers has been traditionally compromised due to the extensive chemical fixation of the tissues. “Among the less utilized clinical samples are formalin-fixed paraffin-embedded tissues,” says Alan Tackett, Ph.D., associate professor, and director UAMS Proteomics Facility at the University of Arkansas for Medical Sciences. “These tissues are extremely valuable as they can be stored long term.”
“We have optimized methods to reverse the fixation of these tissues and have used our approach to perform the most comprehensive study of formalin-fixed paraffin-embedded patient melanoma tissues,” Dr. Tackett explains. He claims that his studies provide the most in-depth approach to uncover biomarkers for diagnosis and prognosis of patient melanoma.
“Our approach for extraction and analysis of proteins from formalin-fixed paraffin-embedded tissues will allow researchers to probe deeper into the proteome of patient samples to try and uncover biomarkers for diseases,” says Dr. Tackett. “Our work will provide researchers with the tools and technological approaches to maximize the analysis of proteins and protein biomarkers from formalin-fixed paraffin-embedded tissues.”
The collection of tissues (up to 35 per donor) and the preservation of RNA posed a unique challenge for the Genotype Tissue Expression Project (GTEx), since all tissue samples were obtained from deceased, low post-mortem interval donors, according to Kristin Ardlie, Ph.D., director, biological samples platform, the Broad Institute.
Dr. Ardlie and her colleagues therefore implemented the PAXgene® tissue system to allow for greater flexibility at both the collection sites and for shipping to the processing and analysis centers. The PAXgene tissue system developed by Qiagen enables histomorphological studies via fixation of tissue samples as well as the preservation of nucleic acids for molecular analysis.
The technique has been shown to enable histological analysis and extraction of high-quality RNA, miRNA, and DNA from single samples. It is also able to preserve intact RNA without crosslinking as well as high-molecular-weight DNA for superior results compared to formalin, according to Dr. Ardlie. PAXgene is capable of preserving high-quality DNA for sensitive downstream applications, including multiplex or long-range PCR, she adds.
However, “given the relative ‘newness’ of the PAXgene system, GTEx has worked closely with Qiagen from its outset to both optimize the collection and preservation of the tissue samples as well as to scale up RNA and DNA isolation from the tissues,” according to Dr. Ardlie.
“Not only has RNA quality been extremely good, but our pathologists working on the project have noted that the histological preservation of the tissues has been as good as or better than that for comparative formalin-fixed tissues,” says Dr. Ardlie.
GTEx is an NIH common fund initiative, launched in 2010, which was designed to investigate human gene expression and regulation in multiple tissues to provide valuable insight into the mechanisms of gene regulation. The project examines individual genetic variation and its correlation with differences in gene expression level to identify regions of the genome that influence how much a gene is expressed and how tissue-specific that expression is.
Dr. Ardlie and Gad Getz, Ph.D., also of the Broad Institute, are co-principal investigators of the Laboratory, Data Analysis, and Coordinating Center (LDACC) for the GTEx project, which also supports tissue and donor data acquisition, and tissue pathology through Cancer Human Biobank (caHUB), as well as multiple centers funded to develop novel statistical methods for the analysis of human gene Expression Quantitative Trait Loci (eQTL).
These groups all work together toward the goal of creating an atlas of human gene expression, and a tissue bank, that researchers can use to study functional mechanisms of inherited susceptibility to disease, clarifies Dr. Ardlie. “The GTEx project is nearing the end of its pilot phase with over 230 donors (>6,500 tissue samples) collected to date,” says Dr. Ardlie. “It now moves in to the scale-up phase where samples will be collected from up to 900 donors.”
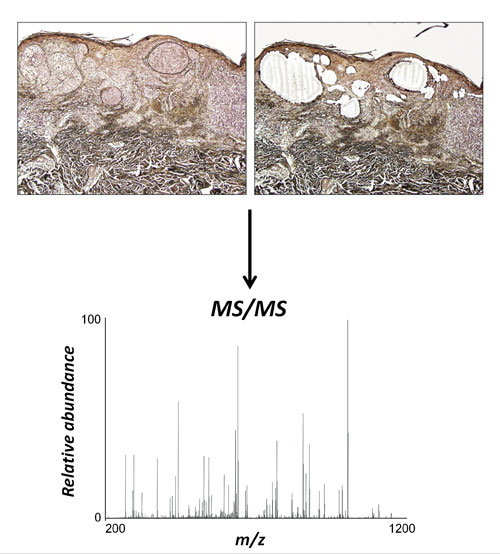
Proteomics of FFPE tissues: Using laser microdissection, specific regions of FFPE samples can be excised for protein extraction. In this case, nests of melanoma cells have been specifically isolated from a skin biopsy. Extracted proteins can then be digested with trypsin and subjected to tandem mass spectrometry. Proteins can be identified by database searching and quantified using label-free proteomic approaches. [Alan Tackett, Ph.D./University of Arkansas]
Direct Digital PCR
Digital PCR (dPCR) is a PCR-based method that allows for absolute quantitation of DNA without the use of a calibrant DNA, as Ross Haynes, a biological science technician at the National Institute of Standards and Technology, explains. “It is a relatively new technology that is anticipated to be widely used in the future.”
Digital PCR enables sensitive and accurate absolute quantitation of a DNA sample obviating the need for a standard curve. A single PCR reaction incorporates many separate reactions, each of which has a positive or negative signal. By appropriate statistical analysis, it is possible to directly calculate the number of DNA molecules in the original sample from the number of positive and negative reactions.
“An understanding of this technology, the potential measurement issues, and platform types will allow labs to make informed decisions about employing digital PCR,” according to Haynes. As he explains, “When you do your extraction, you always end up losing some of your sample, often 80%, resulting in a lot less sample at the end of the day.” The need of the hour, therefore, is absolute extraction efficiency rather than a relative extraction step. Direct PCR methods bypass extraction steps entirely, theoretically allowing the analysis of the entire sample, with fewer manipulations.
“We are looking at direct digital PCR as a way to extract more purified samples, to validate that the technique works with all cells.” Haynes uses trypsinized cells to start with, loading them onto a digital array for image analysis and digital signal processing. The goal is to process clinically relevant samples at the single-cell level, according to Haynes. “PCR buffers and enzymes are resistant to inhibitors, so if we can get PCR to work with whole blood or serum, without having to undergo DNA extraction steps or lose the nucleic acid in the process, that would be ideal.”
Digital PCR is less sensitive than quantitative polymerase chain reaction (qPCR) to PCR inhibitors. The technology is being used at NIST to certify Standard Reference Materials (SRMs) for concentration in copies per microliter. As well, the technology has led to an increased interest in clinical diagnostics based on multiple commercial dPCR platforms. For instance, dPCR has proven superior to traditional qPCR in the detection of low viral loads and low-abundance mutants, Haynes notes.
Selective Enrichment of Mitochondrial DNA
It is now possible to selectively enrich for mitochondrial DNA (mtDNA) from a genomic DNA sample. Eileen Dimalanta, Ph.D., group leader, applications and product development, New England Biolabs, and her colleagues have developed a unique approach for the separation of mtDNA from human genomic DNA, without enriching for highly similar sequences known as NUMTs (nuclear mitochondrial sequences). This is critical for downstream applications such as next-generation sequencing. This novel method relies on the differential methylation patterns of nuclear DNA as compared to mtDNA.
According to Dr. Dimalanta, the simple, cost-effective method can be used to analyze low-level mtDNA mutations from a variety of clinical samples. Furthermore, this technique can be performed using established next-gen sequencing platforms, as well as newer single molecule sequencing technologies.
“Current methods for the detection of mutations in mtDNA involve long-range PCR or hybridization-based capture, followed by next-gen sequencing. These methods lack the ability to distinguish true mtDNA sequences from homologous well-characterized mtDNA sequences in the NUMTs. Co-amplification or co-enrichment of NUMTs complicates the analysis of low-level heteroplastic abnormalities.”
MDx: The Story of Two Approaches
Genotypic tropism tests are considered an alternative to phenotypic testing with similar discriminating ability. The HIV-1 Tropism with Reflex to Ultradeep Sequencing (UDS) based on the genotypic-tropism testing technique is available to physicians in the U.S. in helping the selection of patients for potential treatment with CCR5 antagonists.
Jamie Platt, Ph.D., CGMBS, scientific director, molecular microbiology-gene sequencing, infectious diseases, Quest Diagnostics Nichols Institute, and colleagues, developed a clinical laboratory-based genotypic tropism test for detection of CCR5-using (R5) or X4 variants in HIV infected individuals. The test utilizes triplicate population sequencing (TPS) followed by UDS for samples classified as R5.
Tropism refers to the ability of certain strains of HIV to use different co-receptors. Tropism plays a key role in infection, pathogenesis, and drug susceptibility. While reverse transcriptase, protease, and integrase inhibitors target viral products, CCR5 antagonists such as the FDA-approved maraviroc target specific components of the human immune system rather than the virus per se.
It makes sense, therefore, to use a tropism test before initiation of CCR5 antagonist therapy in HIV-1 infected individuals, as these agents are not effective in patients harboring CXCR4 (X4) co-receptor-using viral variants. UDS had greater sensitivity than TPS to detect minority non-R5 variants. A combination of assays including UDS and TPS showed improved performance compared with TPS alone. The TPS entails genotyping the third variable (V3) loop, a region of the virus that binds to the CCR5 or CXCR4 co-receptor.
Combined with bioinformatic tools, researchers are able to determine tropism in patients carrying R5, X4, or dual-mixed virus. While TPS only detected R5 virus, the highly sensitive UDS is able to detect minority X4 HIV-1 variants, says Dr. Platt. Such an approach ensures that patients with low levels of X4 virus are excluded from receiving CCR5 antagonist therapy.
The key to successful molecular diagnostics, therefore, depends on distinguishing the right gene sequences from the wrong ones. The innovative tools and technologies in nucleic acid sample preparation enriching the regions of interest in the genome may have significant implications in the screening of mitochondrial disorders, HIV tropism, or other viral infections, the analysis of human gene eQTL, and the study of the molecular mechanisms underlying inherited susceptibility to disease.



