December 1, 2012 (Vol. 32, No. 21)
Systems biology covers a broad range of experimental systems. People who study systems biology share the common goal of trying to understand the interactions of many, interdependent factors, and how these interactions govern processes crucial to the organisms’ survival.
Martine Jaworski, M.D., a consulting physician at the University of Ottawa Health Services, believes in using systems biology as a framework for studying diseases, beginning with proteomics as an entry point in the framework.
“Systems biology can be used to connect the omics, from genomics to connectomics,” she said. “Looking at one genetically complex disease in detail, juvenile myoclonic epilepsy or JME, offers an opportunity to test this approach. Systems biology predicts that a large number of proteins would be implicated in JME.”
Dr. Jaworski’s team focuses on ionotropic GABAergic neurotransmission in JME, and the number of proteins implicated in the disease totals more than a dozen.
According to Dr. Jaworski, anatomic networks in animals can be connected to clinical studies in humans, if highly conserved networks or pathways are used.
“The motor pathways are one example, because they are similar in rodents, primates, and humans,” she said. “Connectomics in vivo in human beings, as revealed by newer and increasingly sophisticated imaging techniques, can complement the classical techniques of post-mortem microscopic and other studies, as well as in vivo and in vitro connectomic studies in animals,” she explained during a recent presentation at the “International Conference on Systems Biology” in Toronto.
Dr. Jaworski suggests that connectomics could prove useful in clinical diagnosis, if combined with other levels of omics, such as genomics, transcriptomics, proteomics, and synaptomics, and that pharmaceutical companies also could benefit from applied systems biology. “It is a comprehensive approach that may be very useful in tackling the problem of complex, multifactorial, genetically heterogenous diseases that have hitherto been resistant to other solutions,” she noted.
Phenotypes and Disease
The recent explosion of information with our newfound ease of DNA sequencing calls into question the function of genes associated with diseases. Do particular genes, or combinations thereof, cause diseases? Sometimes this is clear, as in sickle-cell anemia, for example, but determining causality is often more murky.
Gabriel Musso, Ph.D., a post-doctoral research fellow in cardiology at Brigham and Women’s Hospital at Harvard Medical School, studies gene-disease associations to find evidence of causality.
“We have taken a systematic look at embryonic zebrafish, a transparent model vertebrate, to predict novel gene-phenotype associations,” he explained.
“Experimentally, we focused on validating predictions of cardiovascular gene-phenotype associations, identifying some of our predictions as being potentially novel cardiovascular disease genes, one of which had been previously implicated in human heart failure through an unknown mechanism.”
Dr. Musso accomplished this in a machine-learning framework using known, published gene-phenotype associations of morpholino (antisense oligonucleotides used to inhibit specific transcripts). He used “patterns for each phenotype in the expression and sequence features of the genes involved, then ultimately prioritized each of over 15,000 zebrafish genes based on their likelihood to cause any given phenotype upon disruption.” This method was effective in that it was able to weight gene features for predicting phenotype.
Dr. Musso used this approach to predict novel roles for genes with no previous known role in cardiovascular disease. The biggest hurdle, though was experimental validation of their predictions.
“We came up with a 5-category score of heart function and applied it blindly to all injections,” reported Dr. Musso. “As a result we were able to show a clear differentiation between test and control morpholinos.”
Ultimately, he hopes that a better understanding of the many genes involved in cardiovascular disease will aid in developing therapeutics.
Another way of looking at gene networks is studied by Andre Fujita, Ph.D., a researcher in Satoru Miyano’s group at the computational science research program at RIKEN. He presented methods of looking at networks according to the processes that generate them, rather than by the individual connections they contain.
The reason for this approach lies in relationships observed between, for example, neuronal diseases and topological changes in brain connectivity, or genetic diseases and changes in gene regulatory networks.
“However, the comparison between healthy versus disease networks cannot be carried out directly by verifying the presence or absence of each interaction, because there are topological differences within healthy people and also within patients,” said Dr. Fujita. “Even people belonging to the same group present different neuronal or genetic topological features in their networks that make them unique.”
So how do you tell which differences are due to the disease, and which are simply normal variation between individuals? Dr. Fujita and colleagues tried to solve this problem by comparing not just the features of the networks, but also whether the networks are generated by the same process, using statistical methods to compare the generative processes.
They found that networks generated by the same random process share similar features, or spectrum distribution, “which allows us to use it as a ‘fingerprint’ for the networks generated by the same generative process,” said Dr. Fujita. “Based on this relationship, we introduced a concept of ‘distance’ between networks.”
They also developed statistical methods to test whether two networks were generated from the same process. Dr. Fujita hopes to make their methods available to scientists and clinicians, and is currently testing their application to different sets of data in collaboration with others.
“One important step in this direction is to apply our methods in increasing sets of data and show its utility and weakness, such that people acquire confidence and understand the methods,” he emphasized.

Researchers at Brigham and Women’s Hospital at Harvard Medical School are studying gene-disease associations to find evidence of causality. They are using embryonic zebrafish to predict novel gene-phenotype associations. A 48-hours post-fertilization wild-type (above) zebrafish embryo and a morpholino-injected embryo (below) with a cardiac defect and obvious pericardial edema are shown.
Connecting Events in Aging Process
The lab of Daniel Gottschling, Ph.D., an investigator at the Fred Hutchinson Cancer Research Center at the University of Washington, uses the simplest eukaryotes to study complex processes using systems biology. They work with budding yeast, which divide asymmetrically into a mother cell and a daughter cell.
Mother cells continue to divide until at some point no more divisions occur. This finite number of divisions is called the replicative lifespan, which Dr. Gottschling studies as a model for the cellular events that govern the aging process. However, their research had been hampered by the difficulty in isolating mother cells.
“The way this has been done historically is by single cell micromanipulation, and you just can’t do a lot of experiments,” said Dr. Gottschling. “But with our new Mother Enrichment Program, we can now.”
This protocol enables them to isolate large numbers of mother cells more easily. “We’re doing large genetic screens, and we’ve been able to do biology on every organelle, and watch as the cells get older,” he continued. “It has sort of opened up a door.”
Dr. Gottschling began using yeast to study cellular events of aging after observing changes in genome instability as a function of the mother cell’s replicative lifespan.
“We discovered that the mitochondria were messed up just before we saw the nuclear genome instability,” he explained. “The mitochondria, when they became dysfunctional, no longer produced something called iron sulfur clusters,” which are co-factors for many types of proteins, including at least four proteins responsible for genome integrity.
This caused double-stranded breaks, nucleotide excision repair, and base excision repair, to all be compromised in the older cells, according to Dr. Gottschling.
Their recent development of the Mother Enrichment Program has jumpstarted their research, screening for genes involved in aging. They’ve overexpressed each of 250 genes known to be important in mitochondrial dysfunction in yeast, to see whether any could delay the onset of aging.
Using the Mother Enrichment Program to age the cells, and labeled mitochondria for detection, they looked at the effects of each individual transformation on the health of the mitochondria.
They discovered that the yeast vacuole (the equivalent of lysosomes in other eukaryotes) was becoming defective before the mitochondria, and the resulting change in pH of the cytoplasm then caused the mitochondria to become defective.
The lysosome is important for storing amino acids, ions, and small molecules, and requires an acidic pH that is maintained by transporters in their membranes. According to Dr. Gottchling’s hypothesis, when the transporters don’t function properly, a subset of amino acids accumulates in the cytoplasm, destroying the membrane potential of the mitochondria that they require to function, so they become defective.
“In this idea of systems biology, we are actually beginning to dissect causal network interactions,” he pointed out. “One part of the network starts to break down, that is the vacuole, and then because of the interconnectedness to the mitochondria, that then starts to pull it down the hole, and then after that, when the mitochondria start to become defective, the nucleus starts to become defective.”
Dr. Gottschling is excited about the possibility that his research could shed some light on human diseases as well, possibly even explaining previously inexplicable observations of diseased tissue.
“We see fragmentation of mitochondria as they get older,” he noted. “And that’s seen in all these human neuronal degenerative diseases like Alzheimer’s. There are also lysosome defects, but they’ve just seen them as correlates, they’ve never understood why. And so we’re kind of excited that this might explain it. That it might be more than correlative.”
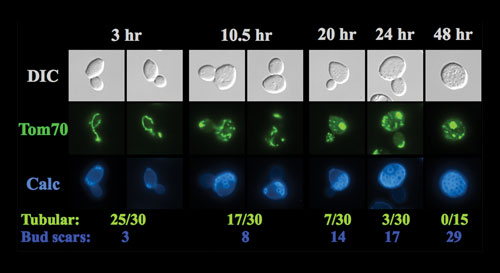
Mitochondria show structural alterations with age in yeast cells isolated using the “Mother Enrichment Program” developed at the Fred Hutchinson Cancer Research Center at the University of Washington.



