June 1, 2009 (Vol. 29, No. 11)
Mass Spec Alternative Detects Biomarker Signatures in Tissues, Blood, and Cell Culture
The current generation of antibody microarray technologies is providing highly sensitive new tools for screening proteins in complex biological samples. Offering an alternative to mass spectrometry (MS) and 2-D gel electrophoresis or chromatography, antibody-based affinity platforms have been developed as high-throughput approaches to the detection of disease-related biomarker signatures in tissues, blood, and cell culture. Last month in Stockholm, researchers revealed their latest microarray-related developments at Select Biosciences’ “Advances in Microarray Technology” meeting.
Providing proof of the commercial viability of truly multiplexed antibody array technologies, SQI Diagnostics’ QuantiSpot™ Rheumatoid Arthritis Assay has been approved in Canada, is CE marked in Europe, and is currently progressing through FDA 510(k) certification. The assay has been developed as a fully automated, highly sensitive, and specific microarray-based fluorescence test for the simultaneous detection and quantification of up to four common rheumatoid arthritis (RA) antibody markers.
SQI reports that the technology represents a major advance in RA diagnosis and monitoring, compared with standard, single-analyte immunoassay technologies and even the more recent advances in multiplexing. “Although multiplex technologies have been developed that can differentiate between rheumatoid factor (RF) and cyclic citrullinated peptide (CCP) antibodies, the systems we are aware of are only capable of demonstrating the presence of antibodies (over a threshold concentration), rather than their concentrations, and don’t provide truly quantitative data,” explained Andrew Morris, SQI’s CFO.
“Clinicians today are interested not only in the presence of anti-RF and anti-CCP but in the concentrations of these subclasses of antibodies, which are increasingly being used to help predict disease progression and make decisions about disease management,” he added.
To address the need for simultaneous quantitation of multiple analytes in multiple samples, SQI’s immunoassay technology has been developed to provide semi-quantitative measurement of anti-CCP IgG, coupled with quantitative measurement of RF-IgA, RF-IgG, and RF-IgM, for up to 76 patients, on an industry-standard 96-well microarray assay plate.
Carried out on the company’s SQiDworks™ platform, the printed CCP peptide and RF antigen microspots capture autoimmune antibodies present in RA patients, and each well includes capture, normalization, and control subarrays. The three internal assay calibrations comprise calibrated concentrations of purified human IgG, IgM, and IgA, to provide an internal dynamic reference standard.
When variations occur, algorithms automatically normalize the capture signal to ensure a comparable signal response across equivalent test spots in different wells and on different plates. Normalized values are then translated to a standardized result.
The technology is fully automated, including sample dilution, incubation, washing, reporter tagging incubation, and final plate conditioning prior to scanning and analysis. SQI maintains the overall process is 80% more efficient than current methods when taking into consideration the four multiplexed analytes. Capable of running up to three plates in a batch analyzing 12 analytes, with a run-time of 270 minutes, the process equates to a throughput of 10.1 patient results per minute.
High signal-to-noise ratios are essential to enable diagnostic-grade precision and accuracy, Morris pointed out. This is in part enabled by high-precision spot printing that binds capture spots to glass substrates, to ensure reproducible arrays of test spots are generated for every well and every plate. This is carried out in the manufacturing process for the kits, which are built at SQI and shipped ready to go.
“Our platform not only represents a fully automated analytical technology for quantitative measurement of antibodies in multiple patients, it importantly includes some 31 confidence tests in each patient well,” he stressed. “Typical ELISA plate technologies may calibrate the device measuring the luminosity signal just once a day. Our technology calibrates for every well, every patient, and every antibody. We essentially overwhelm the test for statistical significance.”
SQI Diagnostics was established about 10 years ago, on the back of technology developed by SQI’s founder and CSO, Peter Lea, Ph.D., for capturing the signal from multiple analytes simultaneously. “The technology is all about generating the best signal-to-noise ratios and getting rid of as much of the background noise as possible. We focused initially on autoimmune diseases, as it was felt the multiplexing technology would be lower risk to develop,” Morris said.
“Development of the RA assay involved optimizing the technology in terms of plate coating, spot detection and analysis, and automation. Now that this has been achieved, we expect development of further tests should be relatively rapid. Our development pipeline includes multiple antibody tests for celiac (6-plex), autoimmune thyroid (3-plex), antiphospholipid syndrome (9-plex), and Crohn’s disease (6-plex).”
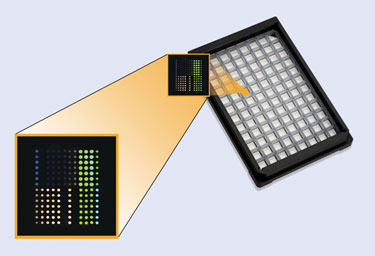
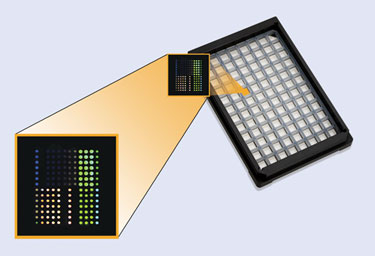
SQI Diagnostics’ QuantiSpot Array can reportedly deliver up to 12 quantitative results in each of 76 patient wells.
Suspension Bead Arrays
Researchers at the KTH-Royal Institute of Technology, Albanova University Centre in Stockholm, have moved away from 2-D planar microarray technology and developed a suspension bead array approach to microtiter-plate-based qualitative proteomics.
Part of the Swedish Human Proteome Resource (HPR) program that generates the Human Protein Atlas resource, the research group, headed by Peter Nilsson, Ph.D., has been set up to develop methods for a systematic exploration of the human plasma proteome using array-based antibody proteomics.
“The technology for using antibodies on color-coded beads is available commercially and by optimizing parameters to ensure reproducibility, high sensitivity, and low background noise, we have developed an approach that can multiplex in two dimensions both in terms of antibodies and samples,” Dr. Nilsson explained. “Assays presently involving up to 96 antibodies profiling 96 samples can be carried out in a single run, detecting proteins down to lower pg/mL levels.”
The main advantage of the approach, in addition to the massive amount of data being generated, is the ability to use whole, labeled samples. “We take plasma samples and carry out direct biotinylation of the whole sample and then, without removing excess biotin, we can interrogate the sample using up to about 100 antibodies. Unlike sandwich assays, which although quantitative, have an upper limit of about 30 antibodies, our technology is only limited by the number of color-coded beads,” Dr. Nilsson said.
The main objective of the HPR center is to produce specific antibodies to human target proteins using a high-throughput production method that involves the cloning and protein expression of protein epitope signature tags. After purification, the antibodies are used to study expression profiles in cells and tissues and for functional analysis of the corresponding proteins in a wide range of platforms. The KTH site in Stockholm is responsible for generating and validating high-quality monospecific antibodies and performing immunofluorescence analyses.
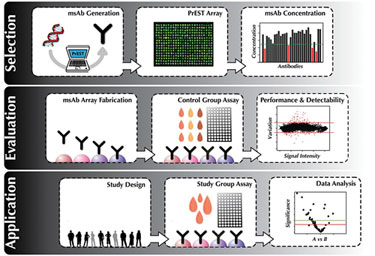
This strategic overview encompasses the selection of produced and validated affinity reagents, over to production and evaluation, and on to screening of dedicated sample cohorts.
(Schwenk, Human Protein Atlas Project)
Fast Screening of Serum Samples
The Stockholm researchers maintain that this bead-based technology offers an alternative to chip-based assays for the fast screening of large numbers of serum samples and has the flexibility of allowing different antibody arrays to be developed on demand, without the need to fabricate new arrays every time.
“We are collecting cohorts from about 25 different diseases,” Dr. Nilsson continued. “The aim is to carry out analysis on 384 samples per month, with 384 antibodies, which corresponds with roughly the number of validated antibodies emerging through HPR research.”
Reverse-Phase Arrays
Researchers at the Natural and Medical Sciences Institute (NMI) at the University of Tuebingen developed an immunoassay technology, known as reverse phase protein microarray (RPPM), for applications including biomarker research, early drug discovery screening, and drug profiling. Based on planar waveguide technology developed at Zeptosens, a Bayer Technology Services company, the approach detects specific protein analytes in hundreds of samples simultaneously using well-characterized antibodies.
NMI maintains that the technology is ideal for investigations in phosphoproteomics (protein phosphorylation), a growing field of research that currently relies on technology involving the enrichment of phosphopeptides or proteins prior to mass spectrometry analysis.
“Drawbacks of the MS approach include the requirement for relatively large samples, time-consuming sample preparation, and problems with automation. In contrast, RPPM technology is significantly higher in throughput, and requires minimal sample and reagents,” explained Markus Templin, Ph.D., head of assay development at NMI. “The technology can be used for the comparative, multiparallel evaluation of several hundred samples, for applications ranging from preclinical toxicity and drug mechanism studies, to protein profiling in healthy versus diseased human tissues.
“It is, we believe, one of the most sensitive fluorescence-based approaches available, as only bound analytes at the surface of the chip generate fluorescence. Signals from unbound molecules in the bulk solution are not detected, which significantly increases the signal-to-noise ratio, allowing the detection of low-abundance proteins,” he added.
RPPM Services Offered
“RPPM assays can be carried out on denatured whole tissue or cell culture samples, and function in virtually the same way as a miniaturized dot blot. The method requires just 500 picoliters of denatured sample, and each spot generated by the arraying instrumentation is 200 microns, which contains about the same amount of material as a single cell,” Dr. Templin reported.
“The technology can detect proteins present at copy numbers down to 600–800 per cell, so even low-abundance proteins, which are increasingly believed to play critical roles in some signaling pathways, can be detected. The array format means we can put up to 320 sample spots onto a single array, six arrays on a chip, and analyze six chips in parallel.
Research at the NMI typically uses up to 250 antibodies, many of which are specific for phosphorylated proteins. At the same time, we use antibodies to detect the amount of total protein, and this allows us to calculate the relative degree of phosphorylation, which corresponds to the overall level of activation,” continued Dr. Templin.
Standardized Immunoassays
The NMI has expertise in antibody development and offers over 100 standardized immunoassays on the RPPM platform. Antibodies for Western blot can also be adapted and validated for the reverse-phase immunoassay technology, according to Dr. Templin.
The institute’s internal research is focused largely on comparing mechanisms in normal and tumor tissues, he explained. “What we tend to find is that while most normal samples have similar signaling pathways, samples originating from different cancers may demonstrate phosphorylation patterns that interplay to different degrees in the different tumor types. The technology is effectively allowing us to compare the signaling pathways that are either common or specific to different tumor types, and compare these with pathways in normal tissue, to get an idea of the mechanisms underlying tumor formation and progression.”
Multiplexed Protein Profiling
In the department of immunotechnology at Lund University in Sweden, associate professor, Christer Wingren, Ph.D., and colleagues have developed a set of recombinant antibody microarray technology platforms for high-throughput, multiplexed protein profiling of crude, nonfractionated samples.
Recently, the team developed the Global Proteome Survey (GPS), for the profiling of complex nonfractionated proteomes. Combining affinity proteomics with mass spec, the technology can probe the whole human proteome in the hunt for disease biomarkers, with a sensitivity sufficient to capture even low-abundance protein analytes down to the picogram/mL range, Dr. Wingren said.
The Lund University technology does not adhere to the one-antibody-one-target approach. “Instead, we have defined a set of peptide motifs of four to six amino acids long, each of which is present in 5–100 different proteins,” Dr. Wingren continued. “The captured antibodies for these motifs, termed context-independent-motif-specific antibodies, are recombinant, single-chain Fv antibody fragments, designed specifically to survive in the assay conditions of a microarray.
“By designing just 200 antibodies that bind to 50 different motifs distributed among different proteins, we can potentially target approximately half of the nonredundant human proteome,” he added.
The overall process involves digesting a biological sample and exposing it to the chip-immobilized antibodies to capture the motif-containing peptides, which can then be enriched and detected using MS. According to Dr. Wingren, such motif-specific antibodies are potentially applicable to any proteome in any species and are not biased toward either abundant proteins or those of particular classes.
The potential utility of GPS led to the establishment of Immunovia by investigators from the department of immunotechnology and CREATE Health, Lund University’s Center for Translational Cancer Research. Immunovia will facilitate the commercialization of human antibodies and biomarkers, primarily for cancer diagnosis and therapy.
“This new technology will be used as a discovery tool in the quest for novel biomarkers, strengthening ongoing biomarker discovery efforts using conventional antibody microarrays,” Dr. Wingren explained. “Our research has already led to the identification of new biomarker signatures, some of which are currently at the validation stage. This is something we believe hasn’t been accomplished using an antibody microarray-based approach before.”