August 1, 2014 (Vol. 34, No. 14)
Clinical genomics soon may be set ablaze by whole-exome and whole-genome sequencing.
Next-generation sequencing (NGS) not only continues to make steady advances in the molecular diagnosis of cancers, it also seems to fit perfectly with our current knowledge of the oncogenome.
In particular, by making it possible to screen the entire coding sequence of cancer-related genes, NGS overcomes a key problem—cancer predisposition cannot be monitored by just a few hotspot mutations.
The sensitivity, speed, and potentially decreased cost per sample make NGS a highly attractive technology. In fact, NGS may soon consolidate many other platforms. While some NGS applications, such as whole-genome sequencing, will probably have to wait several years before they spread and enter routine clinical use, other NGS applications are ready now.
One NGS application that shows immediate promise for clinical oncology testing is exome sequencing. “We hope that in the near future, sequencing of the cancer exome will soon provide oncologists with information they need to identify and utilize treatment options based on the patient’s genomic profile,” says Helen Fernandes, Ph.D., associate professor of pathology and laboratory medicine, Weill Cornell Medical College. Dr. Fernandes, however, adds that NGS will become established in the clinic only after certain challenges are addressed.

Exome sequencing has promise in oncology testing, particularly in the monitoring of disease progression and the prediction of therapeutic responses. To realize this promise, researchers are developing and validating assays of ever-wider scope. [Zmeel/iStock]
NGS Assays
“We need to understand the role of several mutations in the pathogenesis and outcomes of tumors before the information can be used for patient care,” explains Dr. Fernandes. “For a start, we focus our efforts on a panel of 50 genes that could provide actionable information to our clinicians.”
This panel, notes Dr. Fernandes, poses a practical difficulty. It must be used with an assay sensitive enough to identify a mutation present in less than 5% of a heterogenous population of tumor cells.
The most common starting material typically comes from core biopsies embedded in paraffin for preservation. “We were able to successfully use this somewhat compromised material to amplify and analyze multiple genes simultaneously,” continues Dr. Fernandes. The test that has been validated in the clinical lab is able to use 10 ng of starting DNA to generate sequences covering 2,800 hotspots in the 50-gene panel.
The power of this approach was demonstrated in a pilot study aiming to identify sequence variants in head and neck squamous cell carcinoma (HNSCC) with and without human papilloma virus (HPV). Surprisingly, tumors with increased HPV viral copy number are associated with better response to chemotherapy and prolonged survival.
Dr. Fernandes’ team utilized NGS to systematically analyze somatic variants in virus-infected and virus-free samples. The data confirmed key differences in signaling pathways between the two cohorts. Notably, the HPV+ cells retained wild-type p53, which is known to promote cell apoptosis in response to chemotherapy.
Several other genes identified in the study look promising as candidate targets for therapy. “In addition to significant insights in the pathogenesis of HNSCC we were also able to identify potential diagnostic markers that could become therapeutic targets,” asserts Dr. Fernandes. “NGS is rapidly becoming an invaluable tool in translational medicine and oncology.”
CLIA Certification
“I believe that NGS has the potential to become a consolidated platform for multiple genotyping approaches,” says Rajyalakshmi Luthra, Ph.D., director of the Molecular Diagnostics Laboratory at the University of Texas MD Anderson Cancer Center. “NGS has capabilities not only to identify substitution mutations and insertions/deletions (indels), but also to find fusion transcripts and copy number changes.” Instead of managing multiple diagnostic platforms, a clinical diagnostic lab may rely on just one—a next-generation sequencer.
Dr. Luthra points out that while the sequencing technology itself is already capable of detecting indels, variant-calling software packages have not yet reached desired consistency. “We continue visual inspection of raw reads until we can be sure that the software is capable of calling indels with high accuracy,” remarks Dr. Luthra.
Nevertheless, the Molecular Diagnostics Laboratory has undertaken a concerted effort to validate several NGS platforms using CLIA (Clinical Laboratory Improvement Amendments) standards. For example, the Molecular Diagnostics Laboratory clinically validated the Ion Torrent PGM (Life Technologies) using 10 ng of DNA from 70 paraffin-embedded solid tumor samples. Variants detected by sequencing 740 mutational hotspots in 46 cancer-related genes in the panel were confirmed by orthogonal conventional platforms such as Sanger sequencing and pyrosequencing.
Overall concordance between NGS and conventional platforms supported routine use of NGS in the CLIA-certified laboratory. The utility of this approach was elegantly applied to screening mutational hotspots in 54 cancer-related genes using MiSeq, Illumina’s NGS platform, as a part of diagnostic workup and treatment plan for patients with acute myeloid leukemia.
“NGS provides for a single detection platform capable of analyzing multiple genes at the same time starting with a fraction of the material required in conventional multigene analysis,” notes Dr. Luthra.
The laboratory rigorously tested several NGS-based protocols by analyzing and comparing the archival samples with known mutations and samples collected prospectively. NGS platforms showed high level of reproducibility and high detection sensitivity. This high sensitivity was useful in detecting the presence of low-level mutations that were missed by less sensitive conventional platforms during followup and relapse.
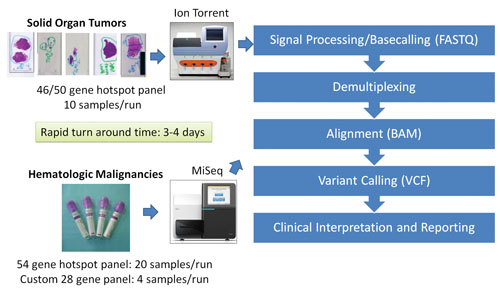
The Molecular Diagnostics Laboratory at the MD Anderson Cancer Center has taken steps to validate several NGS platforms, including the Life Technologies Ion Torrent PGM platform and the Illumina MiSeq platform. The laboratory rigorously tested several NGS-based protocols by analyzing and comparing the archival samples with known mutations and samples collected prospectively.
Universal Oncology Test System
“As Dr. Luthra demonstrated in her study, MiSeq is highly applicable for molecular testing of oncogenes,” says Jennifer Stone, Ph.D., senior market manager for oncology, Illumina. The company has demonstrated conformity with the In Vitro Diagnostic Directive, and it has applied the CE mark to the MiSeqDx™, making it the first and so far only sequencer cleared in Europe and by the FDA for clinical diagnostic applications.
MiSeqDx will play an important role in the partnership forming between Illumina and the pharmaceutical industry. The goal of the partnership is to develop a universal oncology test system.
“The current paradigm, [which emphasizes] single-analyte companion diagnostics, is changing,” continues Dr. Stone. “Tumor genetics is highly heterogeneous, and it changes over time with disease progression. The ability to monitor multiple analytes simultaneously and continuously would enable us to manage cancer as a chronic disease.”
Illumina has already established itself as a leader in supplying NGS-based oncology tests for clinical research, with anywhere between 26 and 94 genes in the panels. A universal oncology test system would incorporate 60–70 actionable cancer genes of specific interest to the pharmaceutical partners.
Many analytes could represent targets of new drugs in the pharmaceutical pipeline. Illumina envisions that such a universal platform would be able to aggregate and systematize results from both academic and drug development efforts. Wide distribution of a universal test would enable broad patient access.
Dr. Stone emphasizes that NGS seems to be a nearly ideal solution for oncology diagnostics because of its ability to profile multiple genes using only a small amount of starting material and to generate results within a short period of time. Continuously collected data would help to monitor the tumor genetic landscape and adjust therapies as needed to keep tumors from evolving.
“The inherent genetic complexity of cancer and the diversity of the pharmaceutical pipeline make it a challenge to pair drugs with patients who may benefit from them,” observes Dr. Stone. “Our multiplexed tool would enable better pairing at both clinical trial and treatment stages.”
Illumina plans to deliver a fully regulatory-compliant solution that will include a universal test; the MiSeqDx; and sample extraction, library prep, and other supporting technologies.
Actionable Physician Reports
“Our goal is to develop actionable sequencing reports for physicians,” says Rong Chen, Ph.D., assistant professor and director of clinical genome sequencing, Icahn School of Medicine at Mount Sinai. “These reports indicate which drugs would most likely benefit a particular patient, which drugs would have toxicities, and which clinical trials might enroll the patient.”
Dr. Chen’s team builds a decision tree for each FDA-approved treatment as well as drugs that have not yet been approved but appear to have potential benefits, as indicated by molecular network information or in vitro or in vivo studies. A sophisticated Hadoop-based search engine combs though vast amounts of publicly available knowledge and integrates information at the DNA, RNA, and protein levels.
“While the bioinformatics workflow to generate driver mutation analysis is very involved, the resulting report should be tailored to the physician’s workflow,” continues Dr. Chen. “We received a lot of feedback from oncologists on how to provide the most useful information and layouts. In the future, we hope to automate the process to reach a critical mass of physicians and then start receiving their feedback regarding success of the suggested treatment.”
Dr. Chen recognizes that genome interpretation may be confounded by inaccuracies that arise from the sequencing process itself or related procedures such as alignment, variant calling, and functional annotation. Dr. Chen’s team deploys multiple strategies to address the known causes of error. These include sequencing on two different platforms and utilizing multiple variant callers and sequence aligners. In addition, the team manually reviews the most important calls and checks raw reads of known driver mutations to avoid false-negative calls. “Mistakes are unacceptable,” insists Dr. Chen. “Treatment outcomes depend on accurate information.”
Dr. Chen’s group continues to develop methods to integrate and translate various molecular measurements in the public repositories into biomarkers for the diagnosis of disease. For example, the group’s ActiVar database contains 110 million distinct variants; 600,000 genome, exome, and genotyping data points; and manually curated human genetics papers.
Dr. Chen’s latest initiative, the Resiliance Project, aims to find secondary mutations that “balance” the disease-causing mutations. It aims to find individuals with catastrophic genetic mutations that somehow remain healthy, presumably via protections afforded by yet undiscovered genetic or environmental factors.
The Resiliance Project is the first systematic attempt to recruit “unexpected heroes” —people willing to donate their DNA in order to further research into mechanisms of disease prevention. “This project takes full advantage of all our previous experience in generating the highest quality sequencing and annotation workflow,” concludes Dr. Chen.
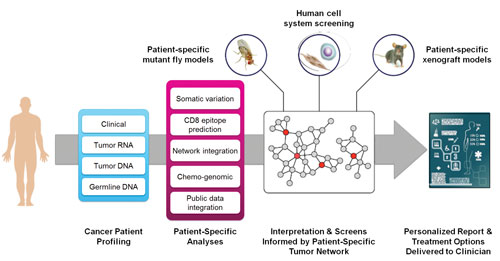
To develop a system capable of generating actionable sequencing reports for oncologists, researchers at the Mt. Sinai School of Medicine are leveraging predictive network models that can comb through vast amounts of publicly available knowledge and integrate information at the DNA, RNA, and protein levels. The researchers also build a decision tree for each FDA-approved treatment as well as drugs that have not yet been approved but appear to have potential benefits.
Overlooked Oncogenes
“Next-generation sequencing has a bright future,” comments Artur Kowalik, Ph.D., head of molecular diagnostics at Holycross Cancer Center in Kielce, Poland. He envisions a time when every newborn is genetically screened for disease predisposition.
According to Dr. Kowalik, enabling technologies that could make mass screening cheaper and faster are already in development—which is not to stay that valuable screening applications don’t already exist. Even currently available benchtop sequencers can make a difference in patients’ lives.
The Holycross Cancer Center has a midsized laboratory that lacks the computational power of large genome centers. “Data analysis and clinical interpretation were our major roadblocks for NGS implementation,” continues Dr. Kowalik. “We had to choose a platform that generated just enough actionable data using commercial software.” A single benchtop sequencer, a panel of 50 genes, and a standardized validated workflow enabled rapid implementation of the NGS technology.
Holycross uses NGS extensively to stratify patients for targeted therapy. Thyroid cancer is a common endocrine malignancy frequently associated with the oncogenic mutation in the BRAF gene (V600E).
“When a patient presents with a suspicious diagnosis for malignancy from fine needle aspiration biopsy of thyroid gland, our endocrinologists routinely call for evaluation of the BRAF (V600E) gene by Sanger sequencing/qPCR,” remarks Dr. Kowalik. In one case, Dr. Kowalik recalls, Sanger sequencing did not reveal mutations in the BRAF. Instead, the Holycross NGS panel was used to uncover oncogenic mutations in a different gene (NRAS). This gene has a positive predictive value for malignancy of about 80%.
Dr. Kowalik gave other examples when NGS assisted in finding mutations that may be overlooked by “gold standard” technologies. In Poland, 70–90% of inherited familial breast cancer results from only three founder mutations in BRCA1/2 genes. Prior to NGS introduction, the diagnostics was based on screening for these mutations only. Now Holycross has an ability to test the whole exome of BRCA1/2 genes.
While the significance of all detected variants may not be yet known, they may still guide the placement of tested individuals into preventive programs. As the analytical capabilities of NGS grow and cost goes down, other family members may be tested for cancer predisposition. “Diagnoses made on the basis of whole-genome sequencing will relieve the uncertainty about breast cancer risk,” concludes Dr. Kowalik.
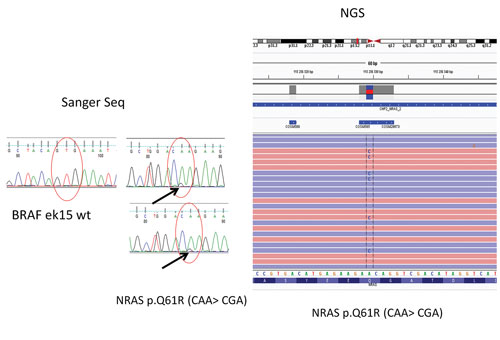
In a case of thyroid cancer that was evaluated at the Holycross Cancer Center in Poland, Sanger sequencing did not reveal mutations in the BRAF (V600E) gene. Instead, an NGS panel was used to uncover oncogenic mutations in a different gene (NRAS). This gene has a positive predictive value for malignancy of about 80%.



