March 1, 2013 (Vol. 33, No. 5)
New Tool Designed to Make It Easier to Work with Soil and Fecal Samples
Metagenomic nucleic acid extractions, protocols in which the entire genomic material of all organisms inclusive of a sample are extracted, are becoming increasingly popular in research areas such as the Earth Microbiome Project and the Human Microbiome Project. Analysis can be done in the context of either fundamental research on microbial communities and biodiversity, or for diagnostics, disease tracking, or biotechnology applications.
Metagenomic extractions typically involve an entire biome lysis, with subsequent nucleic acid isolation and purification from complex, mechanically diverse, and thus difficult-to-lyse matrices. The most common starting materials in metagenomic extractions are soil and fecal samples; these samples are the most difficult to standardize in terms of processing procedures.
Challenges in processing result from variables such as complex matrices with a wide range of mechanical and rheological properties and the inhabitance of innate PCR inhibitors and degrading enzymes. Additionally, the large diversity of cells, tissue remains, and microorganisms produce genomic extractions of varying nucleic acid length and molecular weight.
Fecal samples come in seven basic forms, as classified per the Bristol Stool Chart (Figure 1), and can range from the turdaceous hard solid matrices (Type 1) to a thixotropic liquid consistency (Type 7).1 Fecal cellular material types range from the soft-shelled, easily lysed E. coli to echinococcal oocytes and other parasitic ova, some of the most difficult samples to lyse, as well as solid food remains.
In recent years, several general protocols for stool homogenization and nucleic acid purification have been devised, and there are several articles where they were compared in depth.2 A common motif for the protocols included mechanical lysis of the stool sample, wherein the complete, quantitative lysis of all of the stool constituents was achieved, with even the most resistant single-cell constituents, such as gram-positive bacterial spores and ova, releasing their nucleic acids into a protective buffer.
Mechanical lysis was followed by pre-washing involving some form of flocculation and/or other separation steps to remove lignic materials and flocculate inhibitors. The final step included standard protocols for nucleic acid extraction, most commonly solid-phase bind-wash-elute procedures.
This report will review one of the more popular protocols, based on the application of mechanical sample lysis followed up with flocculation and high-capacity liquid silica slurry binding and solid-phase extraction, the combination of which provides the highest yield and purity of the DNA from all types of fecal materials.
Soil samples present equally difficult challenges in sample preparation and nucleic acid extraction due to many of the same variables enumerated in this article, including complexity of matrix types as well as diversity of biological materials contained, and can also have added natural inhibitors in the form of hard mineral matrices and different content and ratios of humics, fulvics, polyphenols, and polysaccharides.
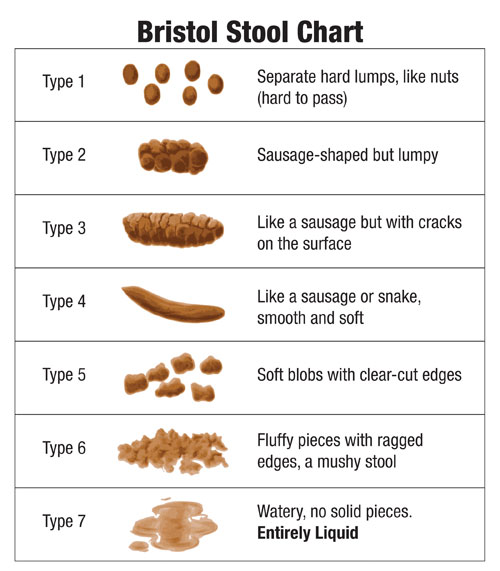
Figure 1. Bristol Stool Chart
FastPrep
In several studies that compared metagenomic sample extraction techniques,3-5 the prevalent protocol for high-yield nucleic acid sample preparation was the FastDNA Spin Kit for Soil™ (MP Biomedicals), based on mechanical lysis utilizing the FastPrep® Systems (MP Biomedicals) with lysing matrix, followed by nucleic acid purification and extraction with either native or modified FastDNA Spin Kit for Soil.
FastPrep-24 and FastPrep-96 are two instruments included as part of the FastPrep system, a leading high-performance mechanical lysis system. These systems consist of disposable, optimized single-use lysing matrix tubes, and a high-performance instrument that creates the 3D nutational orbits with abrupt stops and directional reversals at the apices.
The FastPrep System disrupts complex sample matrices, including tissues and cells, through the multidirectional, simultaneous beating of specialized lysing matrix beads on the sample material, which results in sample breakdown by a simultaneous combination of cascade impaction, mechanical shearing, and fluid flow vortex shearing.2

FastPrep family of instruments and kits.
The lysing matrix is available in standard laboratory formats ranging from 96-well plates and 2 mL tubes, and up to 250 mL bottles, predispensed with optimized combinations of biologically and chemically inert lysing matrix particles. The lysing matrices are application specific; their composition is optimized to provide fast and quantitative lysis of a selected sample type, in most cases within 40 seconds or less. The lysing particles are advanced inorganic materials of different hardness values, sizes, shapes, and densities; the larger beads play the role of cascading impactor and the smaller, sharper particles perform lysis by shearing.
For the purpose of lysing fecal and soil samples, we utilize Lysing Matrix E, which consists of mechanochemically activated silica-based ceramics and glass beads which range in diameter from 0.1 to 1.4 mm, and include larger cascade impaction glass beads of 4 mm. The lysing matrix tubes are fully contained, nuclease-free and disposable, thus preventing cross-contamination.

Content of the lysing matrix E
An overview of the lysing protocol is provided in Figure 2. A sample of the fecal material is placed in the Lysing Matrix E tube, loaded with buffer. Due to the high-throughput nature of the process, samples may be taken from a variety of different geometric positions from the same feces, in order to increase the likelihood of finding the ova6 without adding significant time to the process. The sample is then processed in a FastPrep-24, or for high-throughput applications, the FastPrep-96, for 40 seconds at speed settings of 6.5 or 6.0.
Processing is followed by centrifugation, flocculation/precleaning of PCR inhibitors, binding to a silica slurry column, and then washing and eluting of the purified nucleic acid. This protocol has been documented to provide the highest yield for deoxyribonucleic acid (DNA), and it is due to the extremely high DNA binding capacity of the GlassMilk™ silica slurry.
Additionally, as binding occurs in the gel phase, the protocol may be very easily scaled up as per a user’s specific needs. Furthermore, the consistently reported highest DNA yields are due to the fact that the FastDNA Spin Kits bind nucleic acids in the homogenous liquid phase of the silica gel, which in general has a much larger overall capacity for DNA adsorption than does pre-imbedded glass filters or polymeric membranes. This explains the effect of higher binding capacity and higher DNA yields produced by the FastPrep System and documented in many reports, including a comparison study of four different DNA extraction kits.3
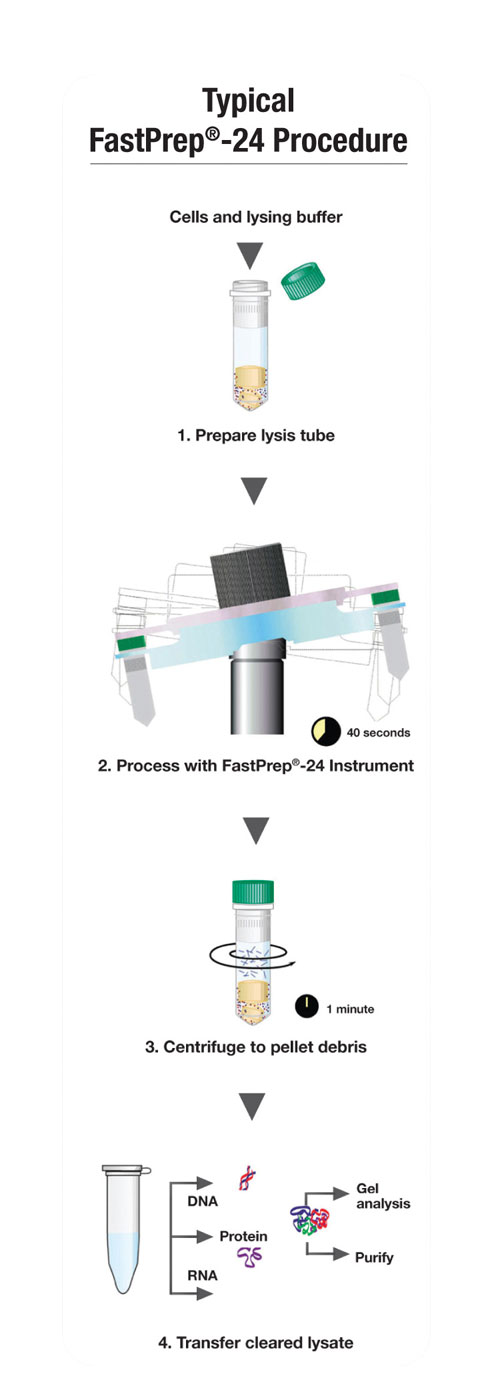
Figure 2. FastDNA workflow overview
The FastPrep System protocol results in a low concentration of the DNA inhibitors after the last purification step, and the entire process can be completed in approximately 30 minutes. In addition to the fast turn-around time, the kits are user friendly in that they are packaged inclusive of all reagents needed for 50 and 100 individual extractions, and they can be easily automated on many of the standard liquid-handling robots/automation stations.
Typical gel from fecal DNA extraction
Conclusions
Recent study of the efficiency of fecal DNA extraction has placed the FastDNA Spin Kit for Soil as the kit with the highest yield of DNA from fecal samples. A study of the pathogen assessment in complex matrices, for example Mycoplasma bovis detection, including feces and feces contaminated soils by Pontirolli et al.,4 has estimated that the FastDNA Spin Kit for soil/feces was the most sensitive method with a volume scalability.
This method resulted in the highest recovery rate, with the minimum load of 4.256 x 106 M. bovis cells per gram of fecal material as a detection threshold, based on the use of a particular PCR primer. In the area of parasite analysis, however, it has been recently demonstrated by Harada et al.,5 that even low concentrations of K. septempunctata, 1.26x 101 spore/g of feces, has been detected by real-time PCR following purification with FastDNA protocols.
This impressive result of essentially one spore per 0.8 grams of feces is done without the optimization of the sample geometric site on the whole sample. Based on recent advances in the understanding of ova and spores spatial distributions,6 it is expected that the detection threshold will be further improved.
Miodrag Micic, Sc.D., Ph.D. ([email protected]), is an instructor of engineering technology at Cerritos College and a vp for R&D at MP Biomedicals. Jeffrey D. Whyte and Veronique Karsten, Ph.D., are global product managers and Liane Miller is director of clinical research and regulatory affairs at MP Biomedicals.
1 Lewis SJ, Heaton KW (1997). Stool form scale as a useful guide to intestinal transit time. Scand. J. Gastroenterol. 32 (9): 920–4.doi:10.3109/00365529709011203
2 Miodrag Micic, Jeffrey D. Whyte (2012). Breaking the Sample-Prep Bottleneck: Automated Lysis Systems Streamline Workflows, Genetic Engineering & Biotechnology News, 32(2): 31-33. doi:10.1089/gen.32.2.16.
3 Merlin W Ariefdjohan, Dennis A Savaiano, Cindy H Nakatsu, (2010). Comparison of DNA extraction kits for PCR-DGGE analysis of human intestinal microbial communities from fecal specimens, Nutrition Journal 9:23, doi: 10.1186/1475-2891-9-23
4 Alessandra Pontiroli, Emma Rachel Travis, Francis Patrick Sweeney, David Porter, William Hugo Gaze, Sam Mason, Victoria Hibberd, Jennifer Holden, Orin Courtenay, Elizabeth Margaret Helen Wellington (2011). Pathogen Quantitation in Complex Matrices: A Multi-Operator Comparison of DNA Extraction Methods with a Novel Assessment of PCR Inhibition, PLoS One 6(3): e17916
5 Tetsuya Harada, Takao Kawai, Michio Jinnai, Takahiro Ohnishi, Yoshiko Sugita-Konishi, and Yuko Kumeda (2012). Detection of Kudoa septempunctata 18S Ribosomal DNA in Patient Fecal Samples from Novel Food-Borne Outbreaks Caused by Consumption of Raw Olive Flounder (Paralichthys olivaceus), Journal of Clinical Microbiology 50: 2964–2968.
6 Stefanie J. Krauth, Jean T. Coulibaly, Stefanie Knopp, Mahamadou Traoré, Eliézer K. N’Goran, Jürg Utzinger (2012). An In-Depth Analysis of a Piece of Shit: Distribution of Schistosoma mansoni and Hookworm Eggs in Human Stool, PLOS Neglected Tropical Diseases, 6(12): e1969.



