September 15, 2009 (Vol. 29, No. 16)
Ability to Dynamically Integrate Intracellular Networks Emerging as Principal Benefit
Metabolomics provides a comprehensive quantitative analysis of small molecules and cellular pathways and a powerful instrument to monitor metabolic perturbations occurring during physiological processes or pathological changes. While just years ago, technical limitations prevented more than one or a few molecules from being explored at one time, the omics revolution facilitated the simultaneous survey of hundreds or thousands of molecules that belong to multiple biological pathways.
Of several approaches used to quantitate thousands of molecules at the same time, separation methods coupled with mass spectrometry have gained widespread use. Nevertheless, while mass spectrometry provides a precise measurement of the molecular mass and can reliably differentiate molecules, it is also accompanied by shortcomings.
“A major challenge is being able to quantify these single molecules,” says Jiri Adamec, Ph.D., lead scientist and director of the proteomics/ metabolomics core facilities at Bindley Bioscience Center, Purdue University. Dr. Adamec will be a speaker at Select Biosciences’ “Advances in Metabolic Profiling” meeting to be held in Barcelona in November.
“In terms of quantification, mass spectrometry is not reporting the overall concentration of the compound but, rather, the ionization efficiency,” he explains. This means that a molecule that does not ionize well might be difficult to measure, despite its high concentrations, while another molecule present at lower concentrations can generate huge signals and be overrepresented during measurements, if it ionizes well.
Research in Dr. Adamec’s laboratory is focusing on modifying molecules with well-defined moieties, for example, the ones containing permanent positive charges that increase ionization efficiency.
A second important challenge for quantification is that ionization efficiency does not depend only on concentration, but is also shaped by surrounding compounds that may suppress ionization, a phenomenon known as the ion-suppression effect. Investigators in Dr. Adamec’s lab solved this problem by differentially isotope labeling two samples and subsequently analyzing them in one run.
With this approach, the relative abundance of the labeled molecules can be measured in a mixture, or when the samples are mixed with a known amount of isotopically labeled internal standards, absolute quantifications can be performed as well. Based on this principle, Dr. Adamec’s lab used in vitro aniline derivatization to quantitate metabolites in the energy and carbon metabolic pathways, and were able to analyze over 35 compounds, including carboxylic acids, phosphorylated sugars, nucleotides, and intermediates belonging to several cellular metabolic pathways.
A current effort in Dr. Adamec’s lab is a targeted analysis focusing on vitamin D derivatization that proposes to develop specific reagents to analyze low concentrations of this vitamin and related metabolites in biological fluids and tissues. This tool would address the low sensitivity, selectivity, and reliability of currently available approaches.
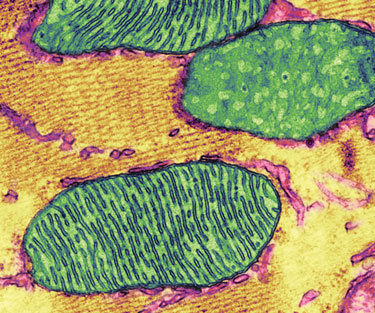
Mitochondrial enzyme errors and mitochondrial fatty acid oxidation defects are responsible for a number of metabolic disorders.
(Thomas Deerinck, NCMIR/Science Photo Library)
Sepsis
Metabolic profiling promises to impact the management of many medical conditions, some of which were never expected to benefit from this approach. At the Barcelona conference, Stephen F. Kingsmore, M.B., C.h.B., B.A.O., president and CEO of the National Center for Genomic Resources, will talk about his work on metabolic biosignatures in sepsis patients. Sepsis, a complex systemic inflammatory response to infection, is diagnosed in 750,000 to 1 million patients in the U.S. annually and is fatal in up to 50% of them.
The clinical presentation of sepsis overlaps with that of noninfectious inflammatory conditions that also present with the so-called systemic inflammatory response syndrome, posing considerable diagnostic challenges, and many markers that are in use such as procalcitonin lack sufficient specificity.
During mass spectrometry analyses performed on the plasma metabolome, Dr. Kingsmore and collaborators described 43 biochemical differences, affecting several metabolic and biochemical pathways, that distinguish sepsis patients from those with nonseptic inflammatory manifestations. Furthermore, the investigators identified discrete metabolic differences, at presentation, between patients who were going to die 28 days later from sepsis and those who were going to survive.
“We found numerous metabolic changes in these patients at presentation in emergency departments that appear to be predictive of death, which is novel and unsuspected,” says Dr. Kingsmore. The ability to use metabolic profiling to guide diagnostic decisions in this patient group has the potential to provide an important screening tool for emergency rooms and underscores the increasing importance of considering metabolomics approaches in the systems biology investigation of complex medical conditions.
“Metabolomics is an area that has a lot of promise, but not a lot is known yet, and there are not yet a lot of thought-informing publications. I think the key need will be for a number of publications to appear that show the value of metabolic profiling,” predicts Dr. Kingsmore.
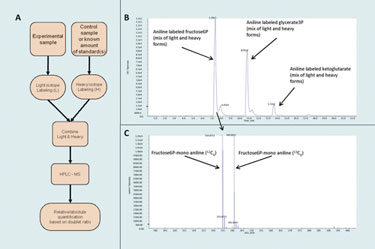
The basic principles of Group Specific Internal Standard Technology (GSIST)
(Purdue University)
UPLC
Also at the Select Biosciences meeting, John Shockcor, Ph.D., director of metabolic profiling business development at Waters, will discuss the impact of reverse-phase ultra performance liquid chromatography (UPLC) on the analysis of lipids from biological samples. Previously, normal-phase chromatography provided certain benefits but the long runs and poor resolution limited its utility for the study of lipids.
“UPLC has brought something that is revolutionary,” says Dr. Shockcor. “We can obtain sharper peaks and resolve large numbers of lipids with little overlap. One of the key features is that these runs are approximately 15 minutes and, in addition, the improved resolution and sensitivity allow a lot more to be performed with the same instrument.” Dr. Shockcor and collaborators have used this approach to collect structural information about specific lipids, which could not have been obtained with any other approaches.
This promises to improve our understanding of metabolic perturbations that occur during complex medical conditions such as diabetes and osteoporosis. While certain people appear to be perfectly healthy for a long time despite increased serum triglyceride or LDL levels, others develop heart disease much earlier and at lower concentrations, and the molecular basis of this phenomenon is insufficiently understood.
“The key to understanding these diseases is to gain a much better insight into lipid metabolism and especially into events that occur early during their progression,” says Dr. Shockcor. “We need to understand lipid biochemistry, and we cannot do that until we have the tools to understand what is going on in depth, within a reasonable time frame.”
Creating Metabolomics Databases
In other recent developments in the metabolomics arena, David S. Wishart, Ph.D., professor in the departments of computing science and biological sciences at University of Alberta, and collaborators have established a fully searchable human metabolome database that currently contains over 6,800 metabolites, ranging from the more abundant (>1 µM) to the more rare ones (
This represents the largest and most comprehensive organism-specific metabolomics database assembled to date and contains chemical, clinical, and molecular information about human metabolites and metabolic enzymes collected from urine, blood, and cerebrospinal fluid samples, according to Dr. Wishart. “The ultimate goal is to identify and quantify as many of the metabolites as we could find,” he adds.
Investigators in Dr. Wishart’s group previously characterized the cerebrospinal fluid metabolome and are currently focusing on establishing a database of metabolites exclusively found in the human serum. The comprehensive inventory of these body fluids provides a valuable resource for investigators from various fields such as biology, medicine, and environmental and plant sciences, who will be able to use the composition lists during their own work.
MetaboAnalyst, a web-based tool developed in Dr. Wishart’s lab, offers several options for data processing and statistical analyses during high-throughput metabolomic studies. “We want to be able to identify the metabolites that we are seeing and to give absolute concentrations. This represents a central challenge and also the key to making metabolomics a true omics science,” explains Dr. Wishart.
Metabologeography
A fascinating new field that became possible thanks to metabolic profiling where other approaches failed is metabologeography, or the differentiation of geography-related metabotypes. Philippe Schmitt-Kopplin, Ph.D., leader of the department of biogeochemistry and analytics at the German Research Center for Environmental Health in Munich, and his French collaborators at the UNSECO Chair Culture et Traditions du Vin and University of Burgundy in Dijon, used a nontargeted metabolomics approach primarily based on ultrahigh-resolution Fourier transform ion cyclotron resonance (FTICR) mass spectrometry to demonstrate that it is possible not only to differentiate red from white wine, but also to determine the type of grapes that were used.
“The challenges in metabolic profiling are the same and are independent of the question you are exploring. The primary challenge is to get an ideal experimental sample setup and to combine the right high-resolution analytical and statistical/mathematical tools,” says Dr. Schmitt-Kopplin.
Using a combination of multidimensional separation techniques to spectrometry (FTICR-MS, 12 Tesla) and spectroscopy (Cryo-NMR, 500 & 800 MHz), his department recently embarked on various studies related to environmental issues (meta-metabolome and C-cycling in the oceans, terrestrial environments, and atmosphere related to global climate change), food chemistry (GMO, beverage metabotyping), nutrition and health (new noninvasive sampling techniques such as exhaled breath condensates and pathogen interactions).
Recently, Dr. Schmitt-Kopplin and collaborators used FTICR mass spectrometry to examine the metabolic profiles of fecal water extracts from twins with Crohn’s disease. Comparing healthy, concordant and discordant twins revealed that there are differences in the metabolome/microbiome, not only between healthy- and Crohn’s-disease individuals but even between patients in whom inflammatory changes were localized primarily in the colon versus ileum.
Besides unveiling new metabolites that are differentially present in patients with the disease and could provide important noninvasive diagnostic or monitoring biomarkers, the approach revealed that not only specific metabolites, but also metabolic pathways, are important to be considered as potential biomarker or therapeutic targets.
Interpretation and Description
The ability to meaningfully interpret information provided by several approaches represents a cornerstone of metabolomics. “We need to integrate all the information, understand how molecules interact with each other, and establish mathematical models that can be validated by experimental approaches,” says Kazuki Saito, Ph.D., group leader of the metabolic function research group at the RIKEN Plant Science Center.
Recently, by using Arabidopsis thaliana as a model, Dr. Saito and collaborators established a link between mitochondrial function and circadian rhythms in plants. Circadian clocks, described in organisms ranging from bacteria to mammals, were implicated in blood-pressure regulation, hormone-level homeostasis, and drug metabolism.
Dr. Saito and collaborators focused on three Arabidopsis genes and, by using transcriptomic and metabolomic analyses, found a link between circadian rhythms, mitochondrial homeostasis and stress response, pointing toward the possibility that mitochondrial physiology and circadian clocks might be linked across several species.
While many omics approaches comprehensively describe various components of the cellular network, the most accurate description of cellular processes is provided by metabolic fluxes, which emerge from the complex interaction of all cellular components.
Fluxomics
Fluxomics is increasingly being shaped as an instrumental tool allowing metabolic phenotypes to be characterized. To address several challenges during 13C-based metabolic flux analyses, Jens O. Krömer, Ph.D., postdoctoral research fellow at the Australian Institute for Bioengineering and Nanotechnology, University of Queensland, and collaborators recently implemented OpenFLUX, as an open-source software allowing investigators to design and interpret metabolic flux studies and circumvent the shortcomings that accompany currently existing approaches.
“The most important application of this tool is in metabolic engineering, where we strongly rely on target identification and need to understand where the bottlenecks are in the network,” says Dr. Krömer. “We also use OpenFLUX to better characterize the metabolism of pathogenic protozoa, ultimately supporting drug development, and this represents a more challenging approach,” he adds.
For applications as diverse as biomarker discovery, drug development, plant engineering, and biofuel production, metabolic profiling promises to substantially impact our ability to characterize cellular processes in their true complexity.
Irrespective of the specific topic, this approach increasingly illustrates how the most complete characterization of biological processes does not emerge simply from a survey of their components, but from understanding their dynamic interplay and the response to perturbations under several distinct conditions.