July 1, 2015 (Vol. 35, No. 13)
Well-Known Technique Can Significantly Improve the Production of Biotherapeutics
The challenges for the industrial production of medicinal products are substantial. For biologics in particular, the process defines the product. As with any process, a series of units of operation must be completed, controlled, and analyzed. For each unit of operation a number of steps exist and at each step a number of possible variables may be present that impact the critical quality attributes (CQA) of the product and, therefore, its clinical safety and efficacy.
Regulatory agencies favor approaches that build quality into a process through the use of Quality-by-Design. However, the number of experiments required to build quality into a process can be vast. In theory it should be possible to test all the possible combinations of variables for a given process, but in practice such undertakings become too large (and expensive) to be performed. Currently the most widely used approach to circumvent this problem is the Design-of-Experiment (DOE) approach.
DOE is a statistical-based tool that reduces the number of experiments needed to model the responses of multifactorial experiments by testing a subset of values for each variable and using those values to predict the impact of changes to one variable on other variables and the product. Ultimately, however, the DOE approach is still limited by the throughput of the experimental method available to analyze the outputs, and it doesn’t test all possible combinations of variables. The amount of experimentation required here should not be underestimated since the challenge for process optimization lies in the combinatorial nature of the problem: how can multiple variables be tested in parallel?
Plasticell’s core technology, CombiCult®, has been designed with that challenge in mind. It is a high-throughput platform that uses combinatorial cell culture technology to screen tens of thousands of protocols in one experiment. The system combines miniaturization of cell culture on microcarriers and a tagging system to allow multiplexing of experiments. Cells grown on microcarrier beads are shuffled randomly, stepwise through multiple conditions using a split-pool method. The iterative process of splitting, culturing, and pooling systematically samples all possible combinations of conditions in a predetermined matrix (Figure 1).
If X number of conditions are tested on each of Y number of cycles, XY protocols are tested simultaneously. Each condition is spiked with a unique fluorescent tag that attaches to the beads. At the end of the process beads bearing cells with the desired phenotype are identified by a screening assay, e.g., immunostaining or reporter gene expression, and individual positive beads are isolated. The culture history of each bead is then deduced by analysis of the tags each one contains.
Protocols are analyzed via bespoke bioinformatics software (AriadneTM), which uses criteria such as hierarchical clustering and probability analysis to select the optimal protocols for further validation. Additionally, Plasticell has shown that it can multiplex different cell lines into each CombiCult screen. This would prove an efficient way to discover protocols that are applicable to many different lines or to investigate differences between cell lines (Figure 2).
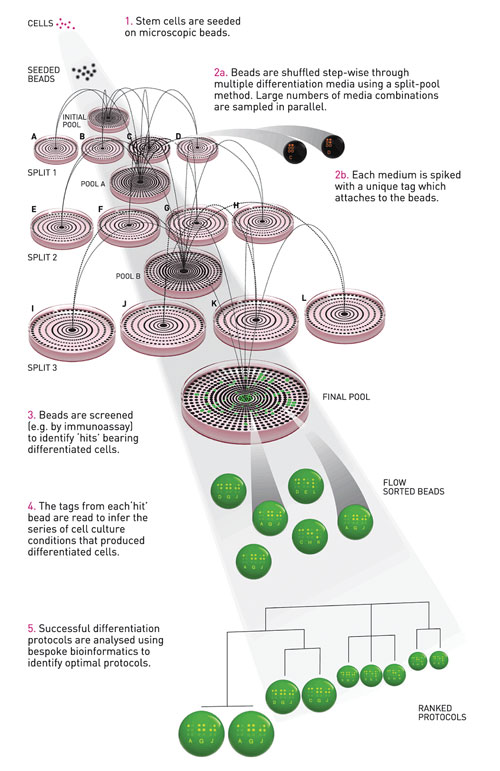
Figure 1. Schematic representation of a CombiCult experiment to find protocols for differentiation of stem cells.
Upstream Processing
How can CombiCult be applied to the upstream process of protein or antibody production?
The efficient and economical production of therapeutic proteins, antibodies, or other biologics, depends on the ability to create cell lines that can synthesize and secrete the protein of interest in high amounts over a reasonable timeframe using processes and materials that are inexpensive and easy to reproduce.
The mammalian expression systems most widely used are CHO cells and hybridomas. These cells are usually genetically engineered to overexpress the protein of interest and to secrete this into the media. Years of research has dramatically improved the yields of proteins from mammalian systems, but there is still room for further enhancement. Optimization of the genetic manipulation of the cells is outside the scope of the technology at present. Nevertheless, clones produced in different ways may be tested simultaneously for other variables.
The first stage in a CombiCult screen is to define the desired cell phenotype and develop an assay to detect it. The most desirable phenotypic characteristic of the cell may, for example, be the protein yield. Protein secretion can be easily detected using FLSS (fluorescent labelling in semi-solid media) if an appropriate antibody for the protein of choice is available.
Using a tool such as Molecular Devices’ ClonePix, beads bearing high-producing cells can be automatically detected by the extent of the halo they produce in the semisolid media containing a fluorescent antibody against the secreted protein, then picked and dispensed in 96-well plates. Further cellular phenotypes can be additionally assessed and detected using immunostaining with other fluorophores.
Once an assay has been established, a matrix with the variables to be tested can be designed. Even though the total number of combinations can be large, there are some practical considerations in defining the number of stages to test and the number of conditions to be tested at each stage. In practice we routinely test between 10 and 20 variables per stage and between three and four stages, making most matrices between 10,000- and 160,000-plex.
For stage 1, for example a number of different clones (say 10) produced using different vectors/cell lines or transduction techniques can be used. These are seeded onto Plasticell microcarrier beads and uniquely tagged, which identifies each individual clone (there is an inbuilt excess of microcarrier beads/pathway, so that if 10,000 possible combinations are tested at least 300,000 beads are seeded). After an overnight incubation to allow cell and tag attachment, the beads undergo first- round pooling and are then split into a further 10–20 conditions where a range of media compositions can be tested.
Each media condition can have either a simple composition with one or two components or a complex one containing a number of small molecules/growth factors/lipids and hormones. Again each condition is individually marked with a unique fluorescent tag that will allow its identification. The cells can be incubated in these conditions for a predetermined period of time and then another round of pooling and splitting can be carried out. During this stage another set of variables can be tested. These can be other media conditions, supplements, or physical parameters such as pH, temperature, and oxygen tension.
At the end of the culture period each pool can be either fixed and immunostained, or plated in semisolid media containing a fluorescently conjugated antibody against the expressed protein. After three to five days, halos of antibody-antigen precipitates can be visualized under epifluorescence, and beads with the largest halos can be picked either manually or using ClonePix.
Once beads are picked, they can be analyzed for tag content and their culture history can be deduced. This data is fed into Ariadne. Depending on the number of positive beads and the degree of clustering a set of 10–20 possible pathways (or sets of combinations) can then be validated, thus reducing the total number of combination of variables from thousands to just a handful.
Mapping the journey of positive beads will define the combination of process steps that best produces the desired cell phenotype, even if the individual process steps are not necessarily the “best” steps. Collectively they will have a better outcome.
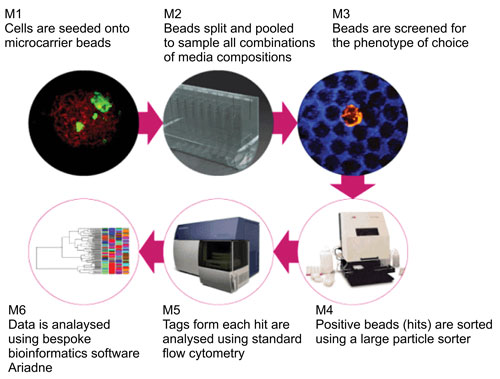
Figure 2. The CombiCult workflow
Diana Hernandez, Ph.D. ([email protected]), is principal scientist at Plasticell.


