June 1, 2013 (Vol. 33, No. 11)
Kinetic Cell-Based Assays Get Into Phase
Time-lapse microscopy is a powerful tool for monitoring phenotypic change, be it cell division, neurite outgrowth, cell migration, or apoptosis. The level of detail and spatio-temporal context this method provides affords significant advantage over most reductionist endpoint techniques. This is particularly true for cellular events that unfold over many hours, days, or weeks. However, capturing and analyzing images over long time periods is technically challenging, especially at a scale to support higher throughput and/or industrialized workflows.
High-content screening (HCS) approaches in microplates—where cells are typically treated, fixed, labeled, and then imaged—have partially alleviated the throughput problem, but largely at the expense of the important kinetic component. Moreover, many cell-labeling techniques can have detrimental effects on cell morphology and function, which can further compromise analyses. To date, little attention has been paid to analyzing label-free phase contrast and/or brightfield images using HCS platforms.
To address these limitations, Essen BioScience has developed a nonperturbing, fully kinetic and quantitative method called Live Content Imaging. This approach, based around the IncuCyte ZOOM™’s “live cell imaging in your incubator” platform, integrates three things: 1) The multiplexed capabilities, image-analysis tools, and quantitative metrics of HCS platforms, 2) The information content of noninvasive time-lapse microscopy, and 3) The versatility, simplicity, and ease-of-use of microplate reader systems.
The IncuCyte ZOOM captures phase and fluorescence (green, red) images from living cells. Simple to use, integrated software modules quantify images on the fly, building real-time, plate-view metrics as the assay time course and biology unfolds. Up to six microplates (96-, 384-well) are housed within a single system, providing sufficient throughput for testing small compound or gene interference libraries.
This approach is widely applicable to adherent cells. The range of exemplified kinetic phenotypic assays includes mono- and co-culture models of cell proliferation, migration, invasion, angiogenesis, neurite outgrowth, apoptosis, and cell death. Measurements of stem cell differentiation, T-cell killing, autophagy, and megakaryocyte production have also been reported.
Some of these measurements are fluorescence-based, and therefore require the use of specialized nonperturbing fluorescent probes suitable for long-term live-cell imaging, such as NucLight™ GFP/RFP. Many, however, are accessible simply through the analysis of phase contrast images. Such are the focus of this article.
High-Definition Phase Contrast Imaging & Processing
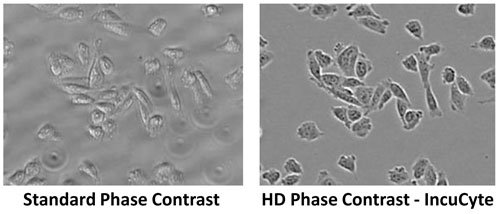
Standard inverted phase contrast imaging in high-density microplates is generally of poor quality because light, which propagates from the annulus ring at the source, is refracted by the fluid meniscus in the well. This causes an aberration that hinders the ability of the objective conjugate ring to remove background light.
The IncuCyte ZOOM overcomes these issues using proprietary methods to yield high-quality phase contrast images in microplate wells without image distortion from the well edge or meniscus of the media (Figure 1). This approach is termed high-definition (HD) phase contrast imaging and presents a significant advance.
In combination with trainable, interactive phase-processing software tools, the IncuCyte’s automated algorithms distinguish cells from background, enabling label-free, phase-based measurements of cell colonies or cell confluence in miniaturized formats.
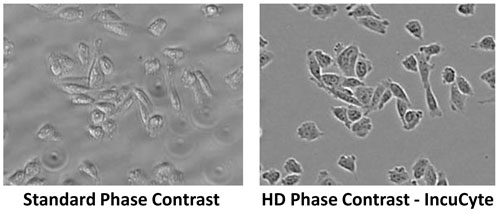
Figure 1. High-definition (HD) phase contrast imaging: IncuCyte’s HD phase contrast imaging corrects for illumination artifacts caused by meniscus and well edges, enabling image acquisition in 96- and 384-well (here shown at 10x) microplates. A standard phase contrast image of the same field of view is shown for comparison.
Once segmented, an image mask is created to display cell-occupied and unoccupied areas, and can be refined using image training sets and filters (Figure 2A). The confluence parameter is often a good surrogate for cell growth. When measured repeatedly over time, it can be used as the basis for kinetic cell-proliferation assays.
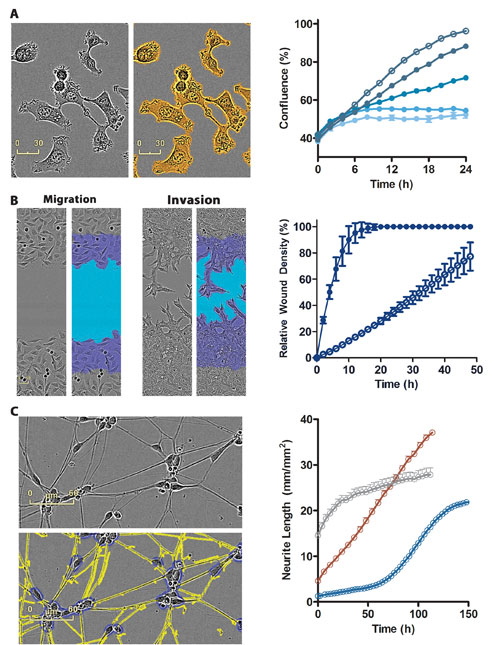
Figure 2. HD phase-contrast images and automated image-processing algorithms are used to quantify complex label-free biology over time. (A) Cell proliferation based on confluence (HT-1080 cells treated with the cytostatic compound, cycloheximide), (B) cell migration (2h) and invasion (36h) through BD Matrigel®, and (C) Neurite Outgrowth (iPSC-derived neurons). In each case, HD Phase images are shown in the absence and presence of the corresponding image mask alongside quantified time course data. In panel B, the scratch wound masks (sky blue) derived from the t=2h (migration) and t=36h (invasion) images are overlaid upon the purple initial scratch wound mask taken from the t=0 image.
Label-Free Cell Migration & Invasion Assays
A similar phase-based algorithm can be applied to in vitro scratch-wound assays, which are commonly used to study signaling pathways involved in cell migration.
Figure 2B shows the image masking and quantification of HT-1080 fibrosarcoma cells on 96-well plates as they migrate across wounds created using an Essen BioScience 96-pin WoundMaker™ tool. HD images were captured and analyzed every two hours for 48 hours and assessed for relative wound density (RWD), a metric based on the comparative density of areas within and outside of the wounded area. With repeat scanning and HD phase image analysis, the full temporal profile of cell migration can be observed.
In this example, the migration of cells is compared to their invasion through a BD Matrigel® biomatrix within the same 96-well plate. Note that 1) The presence of the biomatrix slows the movement of the cells into the wounded area, and 2) The strikingly different cell morphologies under the two conditions with evidence of a mesenchymal phenotype and single-cell invasion tracts in the biomatrix.
Kinetic Measures of Neurite Dynamics
HD imaging also enables the visualization and quantification of fine morphological structures, such as neurites in microplate wells (96, 384), without the need for fluorescent labels and wash protocols. Figure 2C shows kinetic measurements of neurite outgrowth in 96-well plates from rat cortical neurons, human iPSC-derived neurons, and the immortalized cell line neuro-2A.
Using a custom neurite outgrowth module, multiple metrics—including total neurite length, branch points and the number of cell body clusters—can be derived and plotted as a function of time.
In the simple example shown, iPSC-derived neurons send out neurites significantly more rapidly than either primary cortical neurons or neuro-2A cells, once plated. In more sophisticated experiments, it is possible to compare the biology of neurite formation and the retraction or destruction of established neurites, as an example of the value of dynamic measurements.
Live Content Imaging offers an innovative way to evaluate kinetic biology. Image-based phenotypic measurements can be made without perturbing labels or wash steps, thereby reducing experimental artifacts, time, and cost. Additionally, the full time course of long-term biological events can be monitored in real time, eliminating the need to define arbitrary assay end-points, and enabling mechanistic understanding of the dynamics of cellular phenotypic change.
Derek J. Trezise, Ph.D. ([email protected]), is the director and European site head for Essen BioScience.