June 1, 2013 (Vol. 33, No. 11)
MaryAnn Labant
Innovations in live-cell imaging are reshaping scientific paradigms, opening the door to new approaches for scientific inquiry and opportunities for predictive modeling. Emerging technologies allow more in-depth molecular and cellular data collection from living cells and tissues.
Technology advances bring new sets of challenges, including the development of stable processing environments, and analyses of vast quantities of data in robust, transferable methodologies. Live-cell imaging was one of the many technology topics discussed at CHI’s High-Content Analysis conference.
Ammasi Periasamy, Ph.D., professor of biology and director, W.M. Keck Center for Cellular Imaging, University of Virginia, discussed what he called the “fundamentally important role” that technology development plays in the advancement of live-cell imaging.
“Gene chips, high-throughput sequencing, and real-time manipulation of macromolecules in living cells have allowed scientists to observe and understand the central dogma that constitute cellular life,” Dr. Periasamy said.
“Super-resolution techniques have demonstrated that the wavelength of light does not have to ultimately define the spatial resolution, and 3D localization has been demonstrated, within limited volumes, in super-resolution technology,” he added.
Further, Dr. Periasamy said that “improvements of key fluorophore properties, particularly greater photostability, larger Stokes shifts, and smaller sizes, will help to improve current probe designs, which will expand the design space to create a new generation of smart probes. Phytochrome, phototropin, and light-sensitive ion channels derived from bacteria will be used to create tools for live-cell imaging and optogenetics in the near-term future.”
However, despite technological developments, even when high-quality image data are collected, the analysis required to obtain robust quantitative information is extremely challenging, as is the assessment of image segmentation and tracking software accuracy, as well as the analysis of very large datasets.
This is an exciting time for biology, according to Anne Plant, Ph.D., chief, biosystems and biomaterials division, National Institute of Standards and Technology (NIST). “Technology has evolved to where we can now probe large numbers of individual cells in the time domain by light microscopy. This capability provides insight into the statistical details of population dynamics, and tells us about the forces that constrain cell response within an environment.
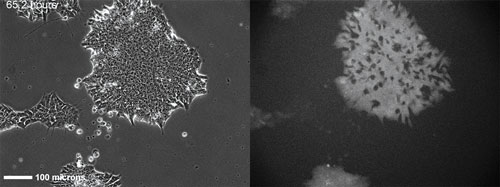
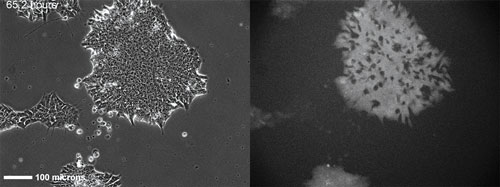
An H9 embryonic stem cell line stably transfected with a GFP-Oct4 promoter construct 65h after seeding and continuous imaging. Time-lapse images over a total of five days allow visualization of how inhomogeneities in colony expression of Oct4 develop over time. Hundreds of colonies are imaged over ~1cm2 every 45 minutes in each experiment. [NIST]
Predictive Model Development
“These dynamic data allow the development of predictive models, and also provide a direct correlation between individual cell response and the fate of that cell,” she continued.
Promoter activity varies from cell to cell, and results in variations in cell fate. In addition, the rate at which promoter activity fluctuates with time in individual cells varies.
The time-dependent fluctuations in expression levels for a transcription factor such as Oct-4 can provide a rate constant that allows for the prediction of future expression levels.
“With stem cells, we also are examining the data for characteristic patterns of expression that might provide predictive information, or correlations, between expression level and the observed phenotype,” Dr. Plant reported.
“We collect time-lapse image data from many colonies, with as much time resolution as we can, for the longest period possible. This allows observation of the events in stem cells that lead to spontaneous differentiation, and the characteristics of colonies that maintain their pluripotent state,” added Michael Halter, research scientist, cell systems science group, NIST. “The goal is to develop tools that will allow routine data collection and analysis at this scale for the stem cell biology community and provide insight into control of stem cell differentiation.”
The researchers used inverted fluorescence wide-field microscopes with broad-spectrum light sources, motorized stages and filters, and environmental chambers. They also developed and evaluated automated segmentation and tracking algorithms. Drs. Plant and Halter then used specialized software tools to visualize very large image datasets.
Drs. Plant and Halter noted that two technologies required to improve the quality of live-cell imaging data are the continuous monitoring of temperature, CO2, and O2 at the bottom of the culture vessel where the cells reside, and the reduction of background fluorescence signal present in cell culture media.
Utilizing the Doppler Effect
David Nolte, Ph.D., professor of physics at Purdue University, also discussed his group’s live-cell imaging work.
“We were investigating a full-frame imaging approach related to optical coherence tomography (OCT) using digital holography. Holography is extremely sensitive to minute motions, and we noticed highly developed speckle fluctuations in the images from living tissues,” Dr. Nolte said. “As we explored these dynamic processes, we came to understand that the origin was in the multiple types of motion occurring at the subcellular scale.”
He noted that “when light shines on things that move, there is a frequency shift, known as the Doppler effect. This is like the effect at a train crossing—the approaching train horn is high-pitched, but as it passes by it becomes low-pitched. When we shine light on living tissues, all the internal motions of the cells cause a similar effect on the light frequency, which we detect and relate back to intracellular motions.”
Tissue dynamic imaging (TDI) is the use of label-free and noninvasive intracellular motion as an imaging contrast agent. TDI provides an opportunity to develop a new 3D tissue screen for many types of drugs in early drug discovery.
Intracellular motions are altered by different mechanisms of action and generate drug-response spectrograms that act as fingerprints for phenotypic profiling. A general technique, TDI can be used for a wide range of applications in biomedicine, drug discovery, biology, and in the clinic. Anywhere cells are aggregating and living, this new form of microscopy can provide unique insights.
“No one knows how to get high-quality information out of living tissue, so they avoid asking those kinds of questions. With TDI, we now provide that needed tool, and we foresee a significant growth in live-tissue imaging,” Dr. Nolte concluded. “TDI relies on light, and we can go about 1 or 2 mm into tissue. This is much deeper than other light-based techniques, but it is not anything like magnetic resonance imaging.”
More Quantitative Information
Fluorescence lifetime-based readouts of protein interactions can provide more quantitative information for fluorescence assays compared to intensity-based readouts. Lifetime-based readouts are independent of probe concentration and of the spectral transmission properties of the instrument, or the sample, and thus can be readily translated between different instruments, or from cells in culture to live disease models.
The Photonics Group at Imperial College London has developed its third prototype of an automated fluorescence lifetime imaging microscopy-Förster resonance energy transfer (FLIM-FRET) multiwell plate reader based on time-gated imaging. They are applying this prototype to probe protein interactions in cell-signaling networks.
“We are also investigating the application of FLIM and FRET to read out biomolecular interactions in live zebrafish, which can serve as disease models, such as for cancer and inflammation. Since they are relatively transparent, they are optically accessible for longitudinal studies. This means that fewer animals are needed to study disease progression,” commented Paul French, Ph.D., professor of physics at Imperial College London.
“Of course, murine studies are still necessary at some stage because their physiology is closer to humans but here the strong scattering and attenuation of optical radiation present challenges for fluorescence assays.”
“Because fluorescence lifetime-based readouts are more robust in the presence of scattering and absorption than intensity-based readouts, they could provide more quantitative in vivo functional information, for example of protein interactions, and longitudinal studies could again reduce the number of animals required for testing. However, it will still be difficult to obtain high-spatial resolution in vivo.”
In the future, Dr. French believes that high-content analysis of 3D cell and tissue cultures could become an important approach, including for FLIM and FRET multiwell plate assays, and could replace live disease models for some studies.

Researchers at Imperial College London are working to provide more quantitative information with noval approaches in fluorescence lifetime imaging on an automated multiwell plate reader. [J. Biophotonics 6(5), 2013.]
Tracking DNA Repair Kinetics
Sylvain Costes, Ph.D., principal scientist in the biocomputational modeling and imaging group at Lawrence Berkeley National Laboratory, spoke about his interest in the dose dependence of the response to ionizing radiation, and the repair kinetics of human cells. “We believe it holds the key to a better understanding of the risk from ionizing radiation,” he said.
“A DNA damage-sensing protein, p53 binding protein (p53BP1) labeled with GFP was monitored as an indirect measure of the incidence of breaks over time. With such an assay, one can monitor the induction and movement of radiation-induced foci (RIF) in live cells, and calculate repair kinetics.
“At first, we would locate and image a few fluorescent cells, take the specimen from the fluorescence microscope to an x-ray machine, expose the cell, and then run back to the microscope, try to find the initial location, and start imaging again.
“Even with the specimens on a warm pack, temperature fluctuations occurred and the first time point achieved was often 10 minutes past exposure. Temperature re-equilibration was required when the specimen was returned to the microscope to limit focal changes due to the warming effect of the media. In addition, large cellular movement made acquisition and foci quantification difficult,” Dr. Costes said.
His team has developed novel RIF counting and cell-tracking algorithms along with custom instrumentation. The researchers also mounted a very small x-ray machine, typically used for material composition detection, directly on the microscope. Aligned with the microscope’s objective, the x-ray device allows irradiation of the specimen on the stage. Microfluidics approached help to keep human cells in eight independent chambers under perfect physiological conditions.
Stage controls are used to look at wells, expose different wells to different doses, and to control the time points.
Software allows time-lapse experiments. The software is paused to run an external routine, in this case, irradiation of the cells and allows pauses for different wells, and different dosage levels, providing a precise kinetic experiment.
The patented technology, including the DNA repair kinetic assay, is in the process of commercialization by Exogen Biotechnology, a company that intends to provide services to track genetic repair capacity.
One of the causes of an acute reaction to radiotherapy is slower than normal DNA repair. Using DNA repair kinetic assays could help identify patients with different radiation sensitivities. The company is also investigating the correlation of breast cancer risk and DNA repair kinetics to determine if DNA repair kinetics can be used as an initial risk indicator to warrant additional genetic testing.
“In live-cell imaging, the biggest problem is that we grow cells in a dish, which is far from the real environment. We are really only looking at one aspect of a much bigger problem,” Dr. Costes explained.
“Microfluidics, combined with tissue engineering to create the complexity of a tissue, will play an important role in the future. Microfluidics is becoming very sophisticated and could provide the means to work with cells while mimicking the micro-environment, the scaffold, that cells live in,” he said. “This is an integral part of the puzzle. With microfluidics we have the tools to design these scaffolds, artificially. They could be used in live-cell imaging in a very elegant way.”