January 1, 2015 (Vol. 35, No. 1)
Mix-and-Read Autophagy and Cell Viability Dyes Enable Simple, Rapid High-Throughput Screening
Over the past decade, autophagy pathway has emerged as an important target in drug discovery. However, current methods to assay autophagy-related responses are not easily adapted to high-throughput screening applications. This tutorial demonstrates the utility of Enzo’s Cyto-ID® Autophagy Dye used in conjunction with IntelliCyt’s MultiCyt Cell Membrane Integrity Dye and iQue® Screener as a platform to detect changes to basal autophagy in cell-based assays.
The autophagy response is profiled in a screening workflow for 14 known effectors of autophagy across 6 different cell lines in a mix-and-read, no wash assay.
Macroautophagy, hereafter referred to as autophagy, is a pathway that delivers a portion of a cell’s cytoplasm to the lysosome for degradation. Turnover of proteins and long-lived organelles by autophagy are a normal part of the homeostatic activities of a cell. During periods of metabolic stress, such as nutrient limitation, autophagy is upregulated to generate the necessary macromolecules for new synthesis.
In addition to its importance in the stress response, autophagy is known to play a role in neurodegenerative, liver, muscle, and cardiac diseases, as well as cancer, aging, and clearance of infections. The involvement of autophagy in a wide variety of disorders has made it an attractive target in drug discovery.
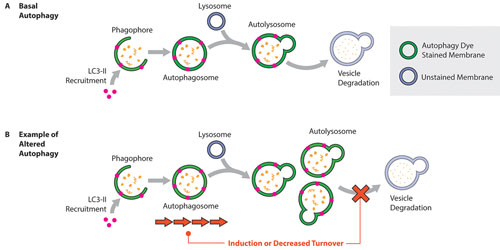
Autophagy initiates with the formation of a phagophore, which is a membrane that begins to surround a portion of the cytoplasm (Figure 1). The phagophore develops into a fully enclosed vesicle called an autophagosome that completely sequesters a portion of the cytoplasm. The autophagosome then docks and fuses with a lysosome, creating an autolysosome, and the contents of the vesicle and the vesicle itself are degraded.
Most methods of autophagy detection visualize autophagy-related intracellular markers. A commonly used marker is LC3-II because of its association with the fully formed autophagosome. Microscopy or western blotting can be used to quantify LC3-II as a measure of autophagosome accumulation or depletion in response to a treatment. Engineered cell lines are often used, in which case an LC3/fluorescent protein fusion is overexpressed within the cell.
These methods require complex assay development, can be cumbersome in high-throughput screenings, and in the case of engineered cell lines, raise questions of physiological disease relevance.
This tutorial describes results obtained from screening 14 known modulators of autophagy. Some compounds cause significant cell toxicity in addition to perturbations in autophagy. The membrane integrity endpoint measurement is utilized as a primary quality control “flag” whereby samples with less than 80% viability are not evaluated for autophagy.
The assay platform offers several advantages for HTS. Following addition of the dye, the assay requires 30 minute incubation. This assay allows screening for a cellular model of choice without requiring an engineered cell line. The multiplexing ability of iQue Screener allows one to obtain readouts of important biological endpoints such as cell membrane integrity.

Figure 1. Cyto-ID Autophagy Dye stains the membranes of phagophores, autophagosomes, and autolysosomes, with minimal staining of lysosomes.
Materials and Methods
Reagents
Autophagy modulating compounds were obtained: 3-Methyladenine, Brefeldin A, Paclitaxel, PI103, Thapsigargin, Wortmannin, Nocodazole, PP242, Tamoxifen, Vinblastine, Rapamycin, Chloroquine, MG132, and Bafilomycin A1.
Cell Culture and Compound Treatment
PC12, U937, A549, H4, HeLa, and HepG2 were all acquired from ATCC and cultured using the ATCC recommended medium and sub-culturing procedures.
A549 experiment
20 μL of A549 cells at a density of 0.5 million/mL were treated with 100 nM Bafilomycin A1 or 20 μM PP242 in a 384-well plate. After 4 hours incubation (37 °C, 5% CO2) the cells were stained with autophagy dye and read on the iQue Screener.
Compound Profiling
All cells (0.5 M/mL) were treated with compounds and concentrations indicated in Figure 2 for 24 hours. Viability was determined by staining with IntelliCyt’s MultiCyt Cell Membrane Integrity reagent. Note that similar determination of cell viability could be conducted with a probe such as 7AAD.
Dose Response Treatments with Chloroquine and PP242
11-point 1:2 serially titrated Chloroquine and PP242 were tested on each of 6 cell lines (0.5 M/mL) with top dose at 100 μM and 20 μM, respectively. Cells were treated for 24 hours before staining for autophagy.
Results and Discussion
The Cyto-ID Autophagy Dye staining response in singleplex was initially evaluated in A549 cells treated with two well-known modulators of autophagy, Bafilomycin A1 and PP242. Bafilomycin A1 is a late-stage autophagy inhibitor, which functions by preventing lysosome acidification and either blocks lysosome fusion with the autophagosome or otherwise prevents acidification of the autolysosome. PP242 is known to induce autophagy through mTOR inhibition, a serine/threonine protease that acts as a repressor of autophagy. Treatment with both autophagy modulators caused changes to autophagy dye staining of cells.
The assay platform described in this tutorial offers unique advantages for high-throughput screening: it is homogeneous, requiring only addition of the dye to your assay and a 30-minute incubation. Importantly, this assay allows screening of a cellular model of your choice without having to engineer a specific cell line. The ability to multiplex and simultaneously detect cell viability in addition to autophagy provides a powerful mechanism for uncovering compound toxicity that could contribute to artifacts in a potential autophagy screen.
A set of 14 compounds with known mechanisms of autophagy modulation were profiled for autophagy and membrane integrity responses in treatments of 6 different cell lines: PC12, U937, A549, H4, HeLa, and HepG2. All 14 compounds caused a response that varied by cell line (Figure 2). A response was considered significant if it differed by greater than 4 standard deviations from the fluorescence of untreated cells.
PP242 was closest to a universal autophagy modulator; it caused increased autophagy dye staining in all tested cell lines except for PC12. However, the results from PP242 treatment of PC12 were flagged due to significant loss of cell viability. Chloroquine caused an increased staining in PC12, U937, H4, and HepG2 cells while causing no change in A549 and HeLa cells.
The profiles for PP242 and Chloroquine were confirmed with dose-response treatments on each cell type. PP242 caused a dose-dependent increase in autophagy staining in all 6 cell types including PC12, which exhibited
Enzo’s Cyto-ID Autophagy Dye offers excellent flexibility for multiplexing with the IntelliCyt’s MultiCyt Cell Membrane Integrity reagent and IntelliCyt iQue Screener. The HTS assay platform is convenient for screening modulators of autophagy and cell health and does not require an engineered cell line. This translates to saving time in screening assay development in a cellular context.
Mix-and-read chemistry of the dyes requires a single addition of both dyes to assay volumes, followed by a short incubation. 384-well plates can be read, without washing, on the iQue Screener in about 25 minutes. The screening workflow outlined in this tutorial represents a significant improvement in the available strategies for autophagy-based drug discovery.
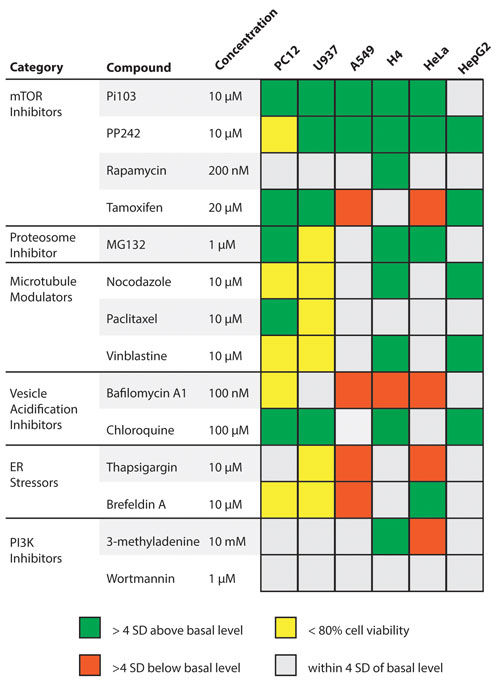
Figure 2. Cellular response profiles for 14 known autophagy-modulators demonstrating increased staining (green boxes) and decreased staining (red boxes).
Kim Luu is currently the director of the assay development group at IntelliCyt. Wini Luty ([email protected]) is a product manager and Arun Kumar is senior technical specialist at Enzo Life Sciences.