December 1, 2007 (Vol. 27, No. 21)
Michael Hoffmann
Jochen Hurlebaus
Christian Weilke
Using the LightCycler 480 System for Discovery and Analysis of Genetic Variation
LightCycler® Real-time PCR Systems from Roche (www.roche.com) are efficient platforms for genetic variation research. They feature the temperature homogeneity and the optical characteristics required for high-performance melting curve analysis. In addition, the plate-based LightCycler 480 System facilitates advanced applications at high resolution in the emerging field of amplicon scanning for new mutations.
Melting curve analysis is a well-established method for characterizing amplicons (e.g., for microbiological identification or for detecting mutations and SNPs). The goal of SNP studies is either the detection of different allelic variants of a given SNP or the identification of polymorphic positions at which previously unknown SNPs occur without determining the allele. Due to the modular design of the LightCycler 480 System, both approaches can be pursued. A broad range of detection chemistries and data analysis algorithms adds flexibility.
Homozygous and heterozygous samples can be differentiated easily using certain saturating DNA-binding dyes that do not interfere with amplification even at high concentrations and give sharp melting profiles (Figure 1). Once SNPs are known, their alleles are best analyzed by melting in the presence of sequence-specific labeled probes that bind with different affinities to different alleles, or allele combinations, in the SNP-containing region (Figure 2).
Melting curve raw data is generally represented by plotting fluorescence over temperature, with the data resolution and temperature range being higher with high-resolution dyes than with labeled probes (Figures 1a, 2a). To make analysis more convenient, the negative first derivatives
(-dF/dT) are often used, revealing melting temperatures at peaks (Figure 1b, 2b).
Data derived from homozygous wild types, homozygous mutants, and heterozygous samples can differ regarding peak number, peak position, or a combination of both. With high-resolution dyes, differences between melting peaks are often detectable but not always large enough to enable clear differentiation of homozygotes (Figure 1b). Using sequence-specific probes allows for a more reliable separation (Figure 2b). The LightCycler 480 Genotyping Software uses the latter approach for identifying SNP alleles.
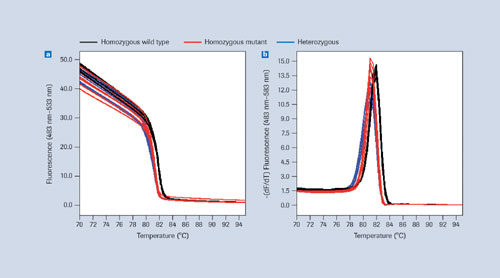
Figure 1
Detection of Sequence Variants
It is not possible to screen amplicons for unknown variations using sequence-specific probes, as the sequence neighboring the potential polymorphic site is unknown by definition. Novel generic DNA dyes can now be used instead, and melting curve data acquired and analyzed at high resolution using LightCycler 480 Gene Scanning reagents and software.
In such an analysis, unknown sequence variations in diploids become apparent in heterozygous samples because they have differently shaped melting curves compared to those derived from homozygous (wild type or mutant) samples. The LightCycler® 480 Instrument is the first plate-based, real-time PCR instrument facilitating such analysis due to its high-performance thermal control and optical components.
LightCycler 480 Gene Scanning Software analyzes these differences by first normalizing the data (see Figure 3a in comparison to Figure 1a) and then temperature-shifting the curves (Figure 3b). By plotting the difference in fluorescence between each sample and its respective wild type, heterozygote samples can be easily identified (Figure 3c). This method has been implemented in LightCycler 480 Gene Scanning Software.
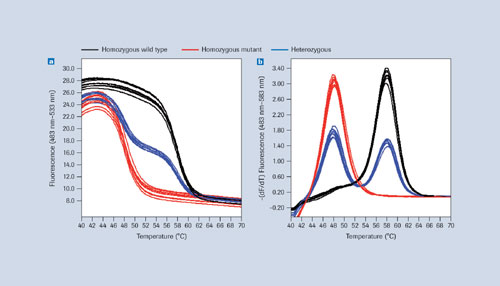
Figure 2
SNP Detection and Analysis Methods
Melting curve analysis is based on a robust post-PCR biophysical measurement and therefore has advantages over other mutation-detection methods that derive information from the amplification process itself. Less sequence data is necessary in order to design a genotyping assay, more information can generally be revealed per reaction, and allele-specific primers or probes are not needed as the same dye or probe can be used for all alleles.
Melting curve analysis with high-resolution dyes offers greater convenience and throughput than traditional methods (e.g., dHPLC) when amplicons are to be screened for the presence of novel genetic variants prior to or instead of sequencing.
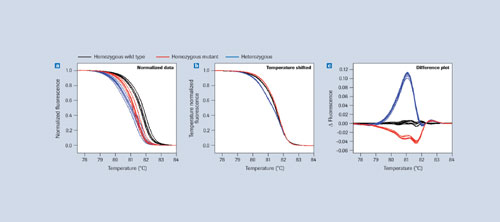
Figure 3
Michael Hoffmann is global marketing manager, Jochen Hurlebaus is manager, innovation concept development, and Christian Weilke is manager, R&D,
at Roche Applied Science.
Web: www.lightcycler480.com.
E-mail: [email protected].







