May 1, 2013 (Vol. 33, No. 9)
RNA Detection Reagent Enables Major Research Step Forward
Detecting gene expression has traditionally been limited to technologies that examine expression in lysed or fixed cell populations. The ability to detect RNAs in individual, live cells can enable an unequivocal assessment of gene expression changes that occur in direct response to specified perturbations. Determining which genes are up- or down-regulated in these perturbed cells provides insight into the relationships between gene expression networks and cell functions.
EMD Millipore’s SmartFlare™ detection reagent is a novel probe capable of detecting specific mRNAs and miRNAs in live, intact cells. This technology allows for carrier-free cellular endocytosis of the reagent, followed by detection and relative quantitative analysis of RNA levels. Because the reagent leaves the cell after the detection event, the same sample can be used for any downstream analysis, meaning you can assess multiple biomarkers or downstream functionalities in the same cells.
Additionally, this reagent requires no up-front sample preparation, has no toxic effects on cellular fate and no known nonspecific, off-target effects. Compared to currently used methods for interrogating RNA that involve examination of non-native, amplified RNA targets, SmartFlare has the potential to provide results that show greater correlation to in vivo observations. In vivo, most cells reside in heterogeneous tissues, and cell-to-cell variation in gene expression can be subtle yet crucial for tissue function. SmartFlare technology can be used to quantitate RNAs in individual cells, providing heretofore unobtainable gene expression information that distinguishes one heterogeneous cell population from another with high resolution.
Figure 1 shows the structure of the SmartFlare probe and its mechanism of action. Each probe consists of a gold nanoparticle conjugated to many copies of the same double-stranded oligonucleotide encoding the target sequence. One strand of the oligonucleotide bears a fluorophore that is quenched by its proximity to the gold core. When the nanoparticle comes into contact with its target, the target RNA displaces the fluorophore. The reporter strand, now unquenched, fluoresces and can be detected using any fluorescence detection platform.
The following general protocol is used for SmartFlare RNA Detection Probes; however, concentrations of probe, incubation times, and detection methods can vary depending on the specific cell types and probes used.
- Culture cells to 60–80% confluency
- Add SmartFlare probe
- Incubate overnight (16 hours)
- Detect via flow cytometry, microscopy, plate reader, or any other fluorescent detection platform
A typical SmartFlare RNA Detection Probe exhibits specificity for its target, as evidenced by the increase in fluorescence upon addition of the target sequence, and lack of signal when a nontarget sequence is added in equal amounts (data not shown).
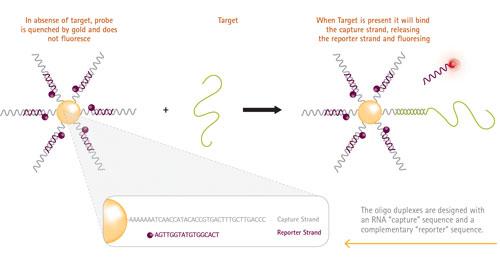
Figure 1. Molecular mechanism of SmartFlare RNA Detection Probe
miRNA Quantitation to Distinguish Cell Types
Because miRNAs are potent regulators of gene expression, frequently determining cell fate, they can be effective biomarkers for disease states. Cell types can frequently be distinguished comparing relative levels of specific miRNAs. A probe to miR-21 was developed to test the ability of SmartFlare technology, in conjunction with flow cytometry, to detect differences in expression between two cell types: HEK-293 cells (expressing low levels of miR-21) and DU-145 cells (expressing high levels of mIR-21). Low expression was indeed seen in the HEK-293 cells (Figure 2, far left section of the histogram) and DU-145 cells showed a distinctly higher expression (Figure 2, far right of the histogram).
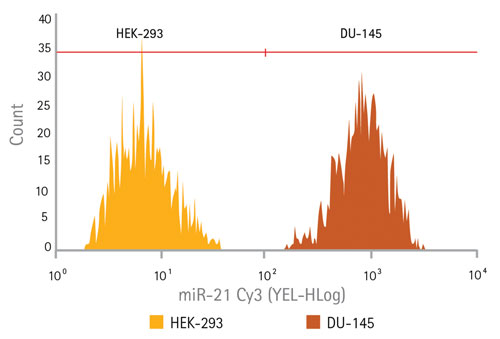
Figure 2. Target-specific miRNA detection using flow cytometry: Detection of a microRNA target miR-21 in both HEK-293 cells (typically used as a low expresser of miR-21) and in DU-145 (a prostate cancer cell line known to express high levels of miR-212) using flow cytometry. The histogram showing the HEK-293 cell population is shown overlaid with the histogram of the DU-145 cell population, reflecting two distinct populations of cells based on their gene expression profile.
Correlates with qPCR Results
By treating cells with SmartFlare probes and measuring subsequent fluorescence with flow cytometry, we measured the levels of two mRNA targets, EGFR and FGF2, in cell types that show differences in expression of each of these targets (Figure 3, left). To confirm expression levels, we quantitated the same RNAs in these cells using reverse transcription PCR (RT-PCR). Because SmartFlare technology allows for downstream analysis following detection and analysis, RT-PCR was performed on the same cells to compare the relative amounts of target RNA in each cell type (Figure 3, right).
As expected, the cells that show lower Ct values correlate to the higher expression by flow cytometry, indicating that the higher level of RNA target present is reflected in the histogram as having a higher mean fluorescence intensity. The histograms revealed additional information on the variation of the expression within each cell population. For example, the HeLa cells (sharp yellow peak) showed much more uniform expression of EGFR than MCF-7 cells (wider orange peak).
Fluorescence microscopy was performed on HeLa and MCF-7 cells following addition of either an EGFR probe or a scrambled sequence. Both cell types showed low levels of fluorescence with scrambled sequence. The HeLa cells showed a dramatic increase in fluorescence with the EGFR probe compared to the MCF-7 cells. The results correlate with our flow cytometry and RT-PCR data (data not shown).
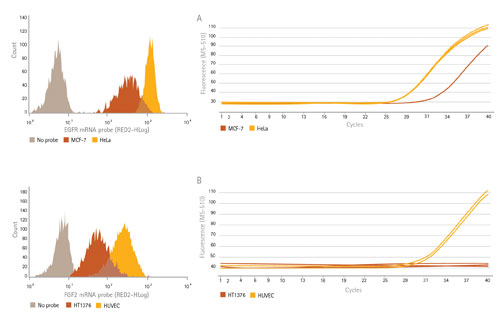
Figure 3. Probe detection of mRNA levels correlates to RT-PCR data. mRNA levels of EGFR RNA (A) in HeLa and MCF-7 cells and FGF2 mRNA (B) in HUVEC and HT1376 cells as quantitated using SmartFlare; both sets of results correlate to their RT-PCR levels. Flow cytometry provides added information at the single-cell level as well as how the expression is distributed within the population.
Conclusions
This novel live-cell RNA detection technology is easy to use and robust. These results demonstrate that the SmartFlare reagent is sensitive and specific—the probe emits fluorescence only upon addition of its complementary target sequence. It can detect both micro and messenger RNA in the cytosol of living cells and has shown good correlation to qRT-PCR. Furthermore the probes are platform agnostic, enabling quantitation via flow cytometry or via microscopy, without amplification.
The many potential uses of this breakthrough technology include sorting of cells based on gene expression (enabling even higher levels of enrichment using additional intracellular markers), live-cell tracking of RNAs, and detection of multiple types of biomolecules (such as protein + RNA) in the same sample. Previous reports have already confirmed that this technology can be used for multiplexed detection of up to three different RNAs (using different fluorophores), enabling the normalization of gene expression levels to “housekeeping” or “control” genes within individual cells.
Notably, SmartFlare technology allows cells to be reused following analysis and after cell sorting for additional downstream experiments on those same cells. By enabling the researcher to discover cells that express particular genes at particular levels in real time, this technology can enhance the significance of observed links between genotype and phenotype in heterogeneous cell populations, truly enhancing the value of data obtained for RNA analysis.
Don Weldon ([email protected]) is R&D manager and Grace Johnston is segment market manager at EMD Millipore.



