March 1, 2008 (Vol. 28, No. 5)
Understanding Interactions Leads to Better Assays as well as Time and Cost Savings
Thorough elucidation of biomolecular interactions is central to understanding biology and disease mechanisms and subsequently devising safe and effective drugs. Life scientists working in academic, pharma, and biotech organizations seek to better understand interactions between protein targets and other proteins or biomolecules as well as the expression of these proteins under different cellular conditions. The kinetics and the affinity of these interactions can affect drug potency and specificity and are relevant to the drug’s mode of delivery, body-clearance rates, and effective as well as tolerated doses.
The FDA has identified protein analysis and assay development as one of the critical bottlenecks slowing the progress of drug development. The demand for quantitative protein analysis has grown exponentially over the last few years as research has identified a large number of putative protein markers that must be monitored under both in vitro and in vivo conditions to establish their true role in disease.
Since 1999, more than 50% of new drugs approved by the FDA have been proteins or molecules that interact with proteins. With increased demand, more and more researchers are challenged with developing de novo protein assays, a task that was previously left to a small group of experts. Previously, tools specifically targeted at accelerating this process were not available. This roadblock to the rapid validation of new discoveries leaves much of the investment in genomic and proteomic research unrealized.
The multiple technologies required for different phases of assay development are slowing progress and increasing costs (Figure 1). The needs of each stage—from reagent qualification and assay development to sample analysis—are unique and of equal importance. These factors highlight the need for a single approach that can be utilized throughout all the stages of assay development. Such a solution to rapidly translate protein discoveries into utilizable assays is now available with the development of the Axela dotLab™ System.
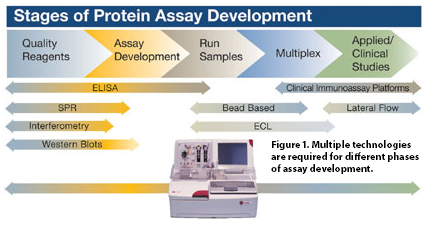
Figure 1
Technology Overview
The dotLab System combines two well-known principles—optical diffraction and affinity reagents—for the quantitative measurement of binding events in real time. Affinity reagents are prepatterned on the surface of a disposable dotLab Sensor to create the diffraction grating.
Assays can employ direct covalent attachment of the target, avidin-biotin coupling, or general protein-recognition reagents. Many assays can be performed label free; however the dotLab System has an extensive sensitivity range and incorporates amplification strategies for low-level analyses.
Existing sensors allow for replicate analysis of single analytes. Sensors under development with user addressable spots allow multiplexing of up to eight analytes in a single sample.
The patterned capture molecules interact with sample in a 10 µL flow channel. The instrument automatically delivers samples and reagents from a standard 96-well plate or bulk containers. Complex matrices including blood, plasma, lysates, and homogenates can normally be used directly without preparation or purification. Sample tubing is integrated with the disposable sensor to minimize risk from plugging or precipitation.
A continuous laser beam interrogates the capture molecules and creates a baseline diffraction intensity. As molecules in the sample bind to the surface, diffraction efficiency is improved in proportion to size and concentration.
A prism molded into the base of the disposable sensor allows operation in total internal reflection mode. This means the measurement beam never passes through the sample, effectively eliminating interference and the need for a separate reference channel.
Binding is determined quantitatively by monitoring the change in intensity of the diffractive signal. The dotLab System displays results graphically in real time. Generation of calibration curves is supported to facilitate the determination of unknowns.
The dotLab technology can be used for a range of applications including the study of biomolecular interactions, protein expression, assay development, and routine immunoassay. The latter two applications as they relate to real-time immunoassays are briefly described in this article.
Development
Quantitative immunoassays are based on interactions between antibodies, the antigen, and other assay components. The process of developing an assay often begins with the qualification of reagents, followed by antibody ranking and selection to identify the best reagents for the application. Antibodies may be produced in animals by inoculation or recombinantly expressed. Additional stages of assay development include identification of the best antibody pairs for sandwich assays, eliminating nonspecific binding and cross-reactivity through the proper choice of reagents, and determining optimal reagent and buffer concentrations.
Real-time measurements of the dotLab System give continuous insight into assay parameters and biological components as they interact in the matrix. This ability to directly observe the relationship between multiple variables can reduce the number of experiments performed by the traditional checkerboard approach by as much as 80%. Development time is reduced from months to weeks resulting in material and labor savings. Candidate antibodies can be screened directly in hybridoma supernatants for both expression and affinity. An example of expedited pairing studies is shown in Figure 2.
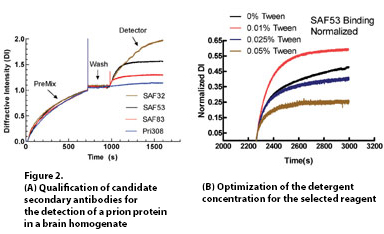
Figure 2
Routine Immunoassay
Capabilities including a wide choice of assay formats, direct detection, quantitative measurement, and an extended dynamic range with real-time data makes the dotLab System ideal for routine immunoassay.
In addition to label-free detection, the ability to run multiple assay formats on a single sample leads to a dynamic range that exceeds 7 logs. Range becomes important in biomarker and diagnostic work where the sample is limited and a panel of analytes must be measured. The system provides an alternative to ELISA and Western blot approaches when a result is needed quickly but also allows automation of multiple samples. Figure 3 illustrates the detection of free PSA, a predictive marker for prostate cancer.
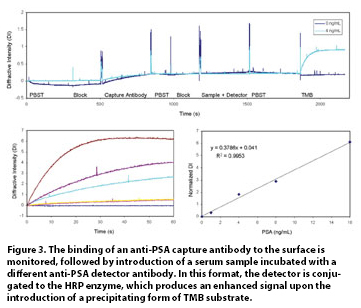
Figure 3
By coupling different affinity reagents to each spot, it becomes possible to measure multiple analytes. The enhanced dynamic range capability can then be applied beyond single-analyte analyses to components of mixtures present at wide concentration differences.
The unique ability to combine detection formats obviates the need to split samples and cluster analytes into similar concentration ranges for separate analysis. There is no observed cross-talk between analytes, a common limitation of multiplexed, end-point assay approaches.
The dotLab System provides a simple, real-time approach for the entire process of assay development and routine immunoassay. Substantial savings in time and money can be achieved by utilizing a single platform across each stage of assay development. In addition, Axela’s dotLab technology allows users the freedom to deploy novel content or use prequalified commercial assays.
Jean-Francois Houle, Ph.D., is
director of technical applications,
and Paul Smith is vp of sales and
business development at Axela.
Web: www.axela.com. Phone: (416) 260-9050. E-mail: [email protected].



