April 15, 2014 (Vol. 34, No. 8)
The cost of bringing a new drug to the market has recently been estimated by Forbes Magazine at $5 billion. This figure derives in large part from high attrition rates in the drug development pipeline that arise from failures in drug efficacy or adverse toxic events, such that in excess of 90% of all small molecules that enter Phase I testing are ultimately not approved.
Collagen Contraction
“The major advantage of using 3D spheroids over 2D monolayer cultures as a drug discovery model is that its geometry and avascular nature mimics the transport barrier of native tissue, where drugs and other molecules experience diffusion limitation, as they get farther away from blood vessels,” said Shiuchi Takayama, Ph.D., professor of biomedical engineering at the University of Michigan.
“The gradients that are established in the cellular microenvironments in the 3D spheroids produce drug response profiles that more closely resemble in vivo responses compared to flat, 2D monolayer cultures,” Dr. Takayama added. Collagen contraction is one of the processes by which cells remodel the surrounding extracellular matrix, which is known to be an important feature of disease progression.
Dr. Takayama’s group has developed a 3D spheroid-based collagen contraction assay that, by miniaturizing the conventional 2D assay, more closely models in vivo diffusion of nutrients and soluble cues. In addition, the assay is more amenable to the high throughput required for efficient drug screening.
Cell-mediated contraction of the collagen matrix is known to be regulated by transforming growth factor-β (TGF-β), which signals via the Smad pathway to modulate the expression of downstream effectors such as connective-tissue growth factor. “We found that brief bursts of TGF-β in the 3D spheroid assay prompted contraction of the matrix, a phenomenon we did not observe in the conventional 2D assay,” remarked Dr. Takayama. Though such modeling of cell-mediated contraction, the 3D assay can more accurately contextualize in vivo biological processes.
Tumor Spheroids
Fredika A. Robertson, Ph.D., executive director for clinical research services at Virginia Commonwealth University’s Center for Clinical and Translational Research, described the development of a tumor spheroid system for small compound screening. This system was developed using tumor cells isolated from the pleural fluid of breast cancer patients.
“These freshly isolated metastatic tumor cells spontaneously form 3D multilayered tumor spheroids, and have several advantages over 2D monolayer cultures for screening,” Dr. Robertson pointed out. These advantages include the enrichment in the 3D spheroids of cancer stem cells or tumor-initiating cells expressing markers such as CD44+, ALDH-1+, and CD133, as well as the recapitulation by the spheroids of the necrotic center that exists in tumors beyond 1–2 mm in size.
“We found that our 3D tumor spheroid system mimics activation of multiple cellular signaling pathways, allowing us to identify the activation of specific signaling pathways in breast metastasis,” said Dr. Robertson. With her group, Dr. Robertson showed that the receptor tyrosine kinase anaplastic lymphoma kinase (ALK) is activated in preclinical models of inflammatory breast cancer, where it signals through a variety of downstream pathways, including JAK1/STAT3, AKT, and mTOR.
“We found that the ALK inhibitor, Crizotinib [Xalkori®, Pfizer] effectively eradicates the 3D tumor spheroids from patients with ALK+ tumors,” Dr. Robertson pointed out. “This is an example of how the identification of the biological signaling pathways that are activated in our patient-derived 3D tumor spheroid systems directly led to clinical trials using this targeted therapeutic.”
Tumor spheroids were also the subject of a presentation by Jason Ekert, Ph.D., senior research scientist at Janssen Research and Development. Dr. Ekert described efforts to develop a tumor model that more faithfully recapitulated the role of cancer signaling pathways in patient tumors.
Pathways involving the transmembrane receptor kinases epidermal growth factor receptor (EGFR) and cMET are involved in the transduction of growth factor signals to the cell, and are thought be active in a large number of lung tumors.
“We were interested in better understanding the differences between 2D monolayer and 3D spheroid lung tumor cultures in the context of the EGFR-cMET cancer biology pathway,” explained Dr. Ekert. “Our flow cytometry data told us that EGFR and cMET expression was reduced in spheroid cultures compared to monolayer cultures, and that basal phosphorylation of EGFR and cMET was higher in spheroid cultures.”
Dr. Ekert suggests that such subtle microenvironment-induced changes in lung tumor cell physiology add a level of complexity to cell-based assays that may be more representative of the in vivo tumor microenvironment, thereby improving the predictive capability of screening assays. He went on to highlight the ability of the 3D system to discriminate between different EGFR and cMET mutants that inhibited cell migration and proliferation.
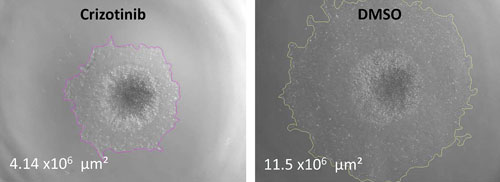
In assays run by Janssen Research and Development, cMET but not EGFR inhibitors reduced cell migration in hepatocyte growth factor (HGF)-stimulated lung tumor spheroids. As shown in these representative images, crizotinib inhibits cell migration pathways in HGF-stimulated lung tumor spheroids. Day four 3D lung spheroids were treated with crizotinib or compound in the presence of 20 ng/mL of HGF for 48 hours to stimulate cell migration pathways. Total area of migrating and spheroid were determined by using bright-field images in a high-throughput Operetta high-content imaging system (PerkinElmer). Magnification: 2× objective.
Co-Culture System
While 3D spheroid systems featured heavily in the talks at the La Jolla meeting, an alternative approach to developing cell models, the organotypic 3D co-culture platform, was described by Ray Mattingly, Ph.D., professor of pharmacology at Wayne State University. “Plexiform neurofibromas (PNs) are present at birth in up to half of Type 1 neurofibromatosis (NF1) patients,” said Dr. Mattingly. “They grow unpredictably to large sizes, have a significant risk of becoming malignant, and have poor five-year survival data.”
A defining feature of PNs is the number of different cell types involved (Schwann cells, fibroblasts, mast cells, and endothelial cells) in which co-dependent cellular growth pathways are activated. “Neurofibromin-deficient Schwann cells secrete stem cell factor that recruits mast cells, which in turn release growth factors and cytokines that stimulate the Schwann cells,” Dr. Mattingly observed.
Such pathway interactions and feedback loops are missing in traditional 2D monotypic culture on plastic dishes, he pointed out. Moreover, the highly active angiogenic pathways in these cells gives rise to significant vascularity that can cause complications during surgery.
“Our goal is to develop the first in vitro organotypic models of NF1 PNs to allow for identification of potential therapeutics,” commented Dr. Mattingly. Our guess is that robustly bioengineered 3D co-culture systems that more accurately model disease pathways and processes will allow preclinical drug screening that is much more representative of likely translational effectiveness than provided by testing in 2D monoculture.”
Development of the co-culture platform is part of the Neurofibromatosis Therapeutic Acceleration Program (NTAP), which is a multi-institutional effort headquartered at Johns Hopkins.
Chip-Based Platform
The transfected enzyme and metabolism chip, or TEAMChip, under development by Solidus Biosciences, is, at its essence, a microarray of immortalized human cells encapsulated in 3D matrices. “The TEAMChip accommodates a diverse array of cells from a variety of tissues, including primary cells, transformed cell lines, and stem cells,” said Jonathan Dordick, Ph.D., co-founder and technical adviser at Solidus and vice president for research at Rensselaer Polytechnic Institute. TEAMChip is based upon 3D cell culture, which employs cells within a physical matrix where the cells are not bound to a surface substrate but interact with the matrix components, such as polysaccharides and proteins.
“Suspension of the cells in the matrix allows them to establish cell-cell interactions that closely mimic the 3D environment within the body, providing a more realistic pathway infrastructure for drug candidate screening for efficacy, selectivity, and toxicity,” Dr. Dordick explained. Cell populations in specific wells of the TEAMChip can be individually transfected with the desired amount of specific gene(s), in effect programming them to express different levels and combinations of molecules that critically impact the response of a cell to a specific compound, such as drug metabolism enzymes, and components of cellular signaling pathways.
“Rapidly identifying the potency, selectivity, and toxicity is a major goal in drug screening,” remarked Dr. Dordick. “Our initial focus with TEAMChip is dialing in the expression levels of enzymes in drug metabolism pathways such as CYP450 and UDP-glucuronyltransferases, which will permit the influence of human metabolism on drug candidate toxicity to be predicted on a more personalized basis.”
Dr. Dordick also highlighted the TEAMChip platform’s potential for transfecting genes within a specific signaling pathway to modulate flux within that pathway. “When drug candidates are screened in such TEAMChips, it is possible to study the interaction of the compound with the relevant signaling pathway,” he concluded.
Using Spheroids for Pathway Analysis
There is increasing evidence that 3D cell culture models are more physiologically relevant, particularly in the context of in vitro tumor modeling, than traditional two-dimensional (2D) cell cultures. Spheroid cultures, self-assembled microscale 3D aggregates of cells, are easy to form, and possess physiological cell-to-cell contacts, secrete their own extracellular matrix, and have nutrient, drug, and oxygen mass-transfer gradients similar to those that exist in vivo.
Combining BioTek’s liquid handling and imaging technologies with the 3D cell culture platform from 3D Biomatrix simplifies generation and imaging of 3D cell cultures for drug-screening applications.
The BioTek MultiFlo™ FX was used to seed, and, subsequently, feed MCF-7 human breast cancer cells expressing GFP and primary human dermal fibroblast cells expressing RFP in the 3D Biomatrix Perfecta3D® 96-well Hanging Drop Plates. The resulting co-culture spheroids were imaged directly in the Hanging Drop Plates using the BioTek Cytation™ 3. Using two-colored co-cultures allows determination of the effects of a cancer-specific drug versus an indeterminate cytotoxic drug.
After five days of spheroid culture, Cyto-ID® Hypoxia Red Detection Reagent (Enzo Life Sciences) was added to the medium in the wells of the Hanging Drop Plate. Following the incubation period, and repeated PBS washes to remove residual hypoxia reagent, 4x imaging was carried out directly in the Hanging Drop Plate using the Cytation 3 to determine if hypoxia could be detected within the different-sized spheroids. Large hypoxic cores were detected in all spheroids, regardless of size.
The hypoxic microenvironment is believed to play an important role in cancer therapy resistance and metastasis, and finding therapies to target hypoxic tumor regions may improve survival rates. Further experiments are planned to determine if inhibiting the hypoxia-inducible cell-surface protein Carbonic Anhydrase-9 (CA IX), in the Hypoxia-Inducible Factor (HIF)-1 pathway will specifically kill cancer cells residing in the hypoxic core.
Spheroid formation is a well-characterized model for 3D culture and screening due to its simplicity, reproducibility, and similarity to physiological tissues. Old techniques were cumbersome and hindered the acceptance of the technology. Today’s methods, make the technique accessible to every life science laboratory.
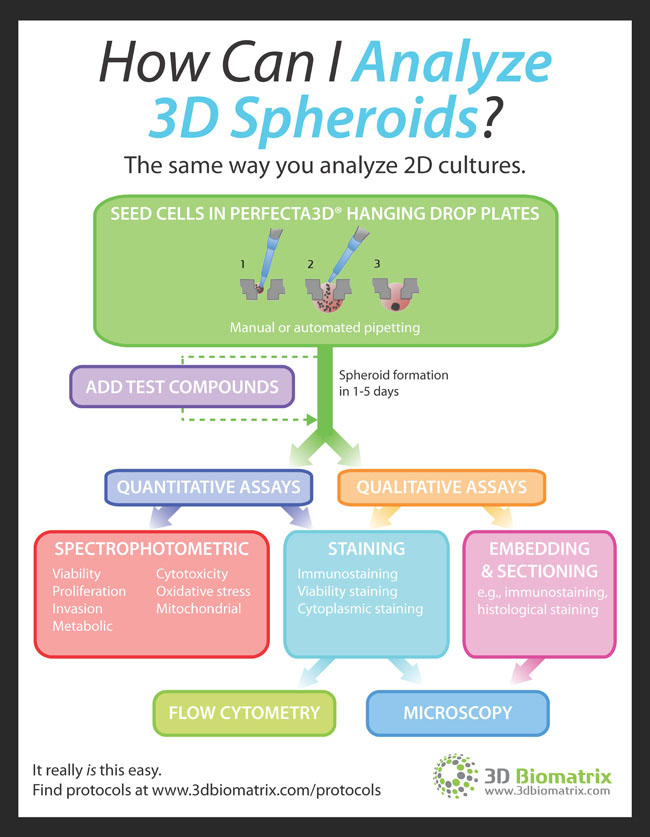
INFOGRAPHIC: Tips on How to Analyze 3D Spheroids
Researchers that are new to 3D cell culture often wonder how they can collect and analyze data from spheroids. The answer is simple: Many of the quantitative and qualitative assays used for 2D cultures can be used for 3D. The infographic to the right from 3D Biomatrix explains everything that you can do with 3D spheroid cultures.