Cancer biomarkers constitute a rapidly expanding research area. Biomarkers represent molecules that could have both diagnostic and therapeutic value in the treatment of cancer and related diseases. Expectations for new products and new therapies are high. An analysis of publication trends using technology landscaping methods provides an objective picture of progress in the cancer biomarkers area.
To construct the technology landscape, we selected promising biomarker candidates based on the scientific literature. For each chosen biomarker, we conducted a separate search in PubMed and retrieved the number of records listed in Table 1 (shown here).
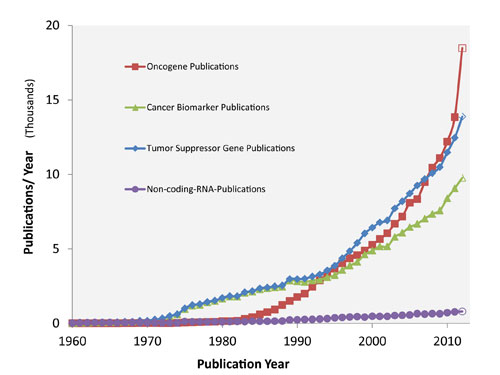
In total, we identified and retrieved 526,185 records from PubMed. The biomarker candidates we selected belonged to four major molecular classes: cancer biomarkers, oncogenes, tumor suppressor genes, and noncoding RNA. Figure 1 presents the growth rate of each publication class. The data show that three biomarker classes (cancer biomarkers, oncogenes, tumor suppressor genes) have been growing at fairly similar rates and that noncoding RNA is growing much more slowly by comparison as it is an emerging and nascent space.
In order to better understand how each biomarker connects to cancer biomarkers/diagnostics, or other diseases, we binned each biomarker search using the descriptive cancer-related keywords listed in Table 2 (which can be found here). From the binned data, we constructed a hotspot map. Figure 2 shows an excerpt segment from this hotspot map. This interesting map illustrates several points:
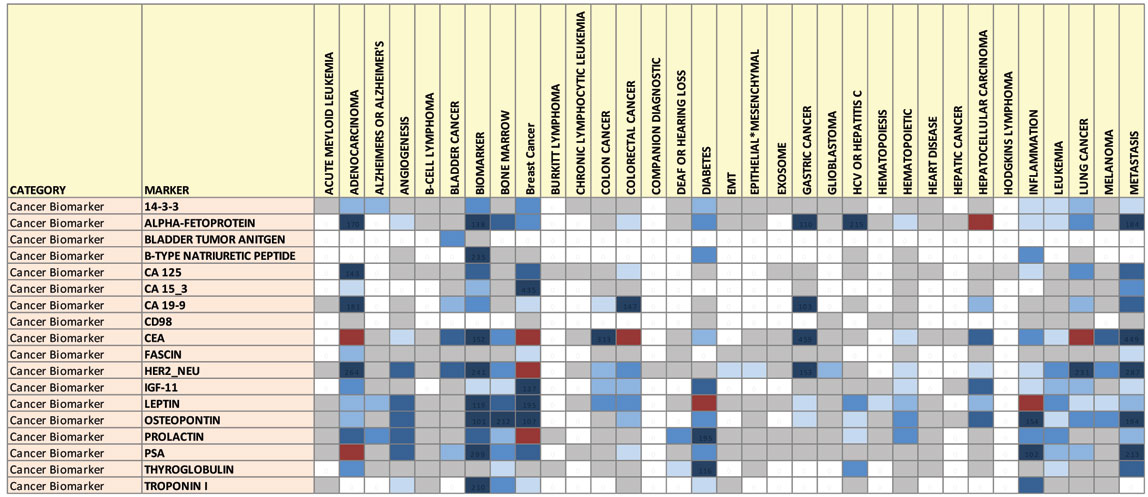
1. Among the biomarkers chosen for study, many biomarkers are connected to several types of cancer.
2. Each type of cancer can be characterized by a unique “fingerprint” of biomarker publication activity. Given the set of biomarkers we selected, the map suggests to us that multiple biomarkers will be needed to form a diagnostic pattern for a given cancer type.
- Some biomarkers (CEA for example) are mentioned in PubMed abstracts for nearly every type of cancer. We believe that this type of biomarker fingerprint is more indicative of the general disease state rather than a specific cancer type
- Others biomarkers (e.g., CD98) are less studied, and the fingerprint is more specific for particular diseases
- Noncoding RNAs are an emerging area and therefore there is a lot of “virgin territory” in this space—representing an area wherein industry participants can pursue a land grab to establish intellectual property positions
3. Hotspot mapping with “fingerprinting” is the start: Validation efforts are necessary to determine if the “fingerprints” are prognostic of the natural course of a disease or are predictive of treatment outcomes.
To our knowledge, this is the first published study that used technology landscaping methods to understand connections between cancer and other disease states and putative biomarkers. Our complete hotspot map allowed us to postulate that specific cross-class biomarker combination signatures may be indicative of specific types of cancer and indeed make predictions of specific “signatures” of putative biomarkers that could be validated using clinical samples from patients.
Only a subset of the complete hotspot table is presented in this article, and we believe that it illustrates the power of studying large datasets of publications to assess not only research trends in these spaces, but also discover and validate new molecular patterns with utility in translational research and biomarker assay development.
Gary Oosta, Ph.D., is strategic technology assessment analyst and blogger (www.emergingtechinsights.com). Enal Razvi, Ph.D. ([email protected]), is biotechnology analyst, Select Biosciences.

