January 15, 2016 (Vol. 36, No. 2)
Joseph M. Beechem Ph.D. Senior Vice President of Research and Development NanoString
Sarah E. Warren Ph.D. Senior Scientist, Immune Oncology NanoString
Simultaneous Digital Counting of DNA, RNA, and Proteins at 800-plex
The tumor microenvironment is a heterogeneous milieu of tumor cells, stroma, infiltrating immune cells, and vasculature, all of which interact to influence the course of disease progression. Cells within this environment respond to constantly changing levels of growth factors, cytokines, oxygen, nutrients, cellular debris, and metabolites.
Cancer cells respond to this challenging environment by constantly mutating and altering their genome, transcriptome, and proteome. Infiltrating immune cells activated by tumor proliferation and associated inflammation can also induce large changes in mRNA and protein expression patterns which have direct consequences on immune-mediated elimination, equilibrium, or escape of the tumor.
Characterizing Complex Interactions
The ability to measure changes to the DNA, RNA, and protein is crucial to understanding tumor responses to changes in the environment or to therapeutic treatment. NanoString® Technologies has pioneered the concept of 3D Biology—the ability to measure any combination of DNA, RNA, and protein simultaneously using a single detector from a small clinical sample—in order to characterize these complex interactions (Figure 1).
The NanoString nCounter® platform directly measures multiple analytes of interest (DNA, RNA, or protein) through simple hybridization reactions, multiplexed together using fluorescent optical barcodes (Geiss et al.). DNA and RNA from the cell or tissue of interest are hybridized directly to nucleic acid-based optical barcodes while proteins can be detected in a similar manner by covalently labeling primary antibodies using small oligonucleotides, which then hybridize to the nucleic acid barcodes (Ullal et al.).
Up to 800 DNA, RNA, or protein targets can be evaluated simultaneously in a single reaction. Only small amounts of starting material are required; sufficient analyte can be obtained from one or two formalin-fixed paraffin sections for DNA/RNA profiling, and/or 150,000 PBMCs for DNA/RNA/protein profiling.
This strategy for barcode labeling and detection of analytes has many advantages over other analytical approaches. First, direct, nonamplified, single molecule digital counting allows for detection over a wide dynamic range with extremely high reproducibility—often over 98% concordance between biological replicate samples. Second, the ability to interrogate DNA, RNA, and protein in a single reaction (as opposed to dividing samples for sequencing, mass spectrometry, cytogenetics, etc.) maximizes the amount of biological information that can be obtained from precious clinical samples.
Finally, the single-molecule detection strategy allows for direct comparisons (without complex normalization procedures) of DNA, RNA, and protein since all analytes are counted digitally using the same detector.
Characterizing DNA, RNA, and protein levels within a sample can greatly enhance the understanding of immune responses to cancer progression. Many key regulatory events are reflected only in the variable expression of mRNA and proteins. For example, macrophages found within the tumor will have identical genotypes, but these cells can be polarized on a gradient of stimulatory (M1) to suppressive (M2) phenotypes.
M1 and M2 macrophages can be discriminated by significantly different expression patterns of activating and inhibitory mRNA and proteins. Post-transcriptional regulation of protein expression by miRNA or other regulatory elements can induce discordance between mRNA and protein abundance, which is not captured by RNA-only profiling.
Furthermore, interpatient variability of the genome can also modulate the M1:M2 balance, e.g., a single nucleotide polymorphism within PTPN22 biases macrophage polarization to an M2 phenotype (Chang et al.). The NanoString 3D Biology approach is designed to measure this type of complexity, greatly increasing the net information-content obtained from small amounts of clinical samples.
Figure 1. NanoString 3D Biology platform enables the discovery of up to 800 DNA, RNA, or protein targets from a single sample using fluorescent nucleic acid barcodes. DNA and RNA are detected by direct hybridization of oligo barcodes, and proteins are detected by barcode conjugated antibodies, which enables measurement of all three analytes from a single sample (1–2 FFPE slides or 150,000 PBMC or dissociated tumor cells) using a common optical reader.
Case Study
As an example of 3D Biology in action, we looked at a model system that reveals the immunological consequences of targeted therapies in the cancer setting. In this study, we simultaneously measured genotype, mRNA, and protein responses in cells treated with the targeted therapy vemurafenib, a BRAF kinase inhibitor specific to the V600E mutation, which frequently occurs in melanoma tumors.
Two cancer cell lines were genotyped using optical barcodes specific for either the wild type or mutant BRAF DNA sequence (Figure 2A). Following treatment with vemurafenib, 770 mRNAs and 30 proteins were digitally counted using the nCounter system. As expected, little change was observed in the BRAFWT cells, whereas the BRAFV600E cells had significant mRNA expression changes (Figure 2B).
Large changes specific to the BRAFV600E cells were also observed at the protein level, where multiple cell surface proteins associated with immune responses were dysregulated in the BRAFV600E cells (Figure 2C). This unexpected change in immunostimulatory potential of the cells was confirmed by reports in the literature of similar findings using Western blots, flow cytometry, and qPCR (Sapkota et al., Wang et al.).
Detection of therapeutic efficacy has historically been focused on the single analyte responses. Targeted therapies were reasonably well served with SNP-based DNA assays to identify responding patients. However, the advent of the immunotherapy revolution demands multi-omics strategies for biomarker discovery and companion diagnostics development because immuno-oncology therapeutics elicit multifaceted changes to the tumor cells and the immune system.
The ability to capture information from DNA, RNA, and protein on a single platform with a high degree of confidence enables a deeper understanding of both the tumor and the associated immune response. Large-scale changes to the mRNA transcriptome within immune cells are one of the hallmarks of an activated immune response. These transcriptional changes lead to an alteration in the cellular proteome, which can determine the success or failure of immunological control of the cancer.
The NanoString barcoding system enables interrogation of any combination of DNA, mRNA, and proteins (up to 800 targets) allowing more thorough characterization of the changes to the tumor and immune system in response to treatment, potentially leading to the identification of novel biomarkers.
Although tumors originating from different tissues show unique molecular and histological phenotypes, immune responses directed against diverse tumors tend to share common features. For example, immune responses at the level of the tumor micro-environment can manifest as “hot”, consisting of extensive tumor immune infiltration and active suppression of tumoricidal activity, or “cold”, characterized by an absence of immune infiltration and lack of an ongoing immune response.
Immune therapies have been shown to be effective against a subset of tumor types, thus creating the potential to generate universal therapeutics for oncology. However, the key molecular events within the tumor microenvironment that predict efficacy remain undefined. The multiplexed, multi-analyte 3D Biology platform has the potential to rapidly enable discovery and validation of a universal diagnostic that can be used by patients and physicians to guide rational treatment decisions.
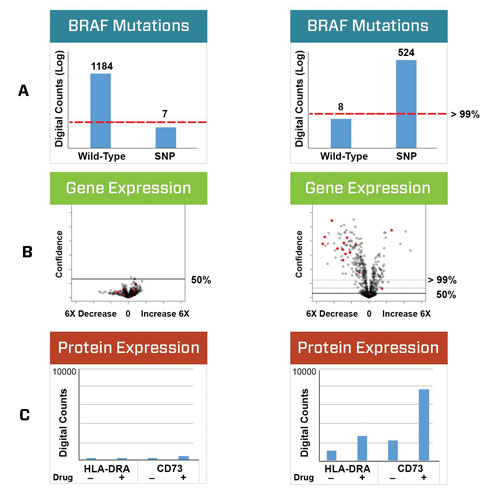
Figure 2. The NanoString assay reveals unpredicted immunological consequences to treatment with BRAF targeted therapy. (A) SW48 and SK-MEL-28 cell lines were genotyped with NanoString optical barcodes specific for WT BRAF or the V600E mutation. The barcodes are able to discriminate the genotypes with >99% confidence. (B) Cells were treated with vemurafenib, a V600E specific BRAF inhibitor, and expression changes were measured for 770 mRNA transcripts using the nCounter Pan Cancer Pathways Panel. Changes in transcript expression were assessed using with NanoString’s nSolver™ analysis software. As expected, vermurafenib treatment only induced expression changes in BRAFV600E expressing cells. Open circles are mRNA, red circles are protein. (C) Treated cells were also analyzed for changes to immune-associated proteins using antibodies from the RNA:Protein PanCancer Immune Profiling Panel. Significant upregulation of immune-associated proteins was observed exclusively in BRAFV600E expressing cells.
References
Chang HH, Miaw SH, Tseng W, Sun YW, Liu CC, Tsao HW, Ho IC. PTPN22 modulates macrophage polarization nadn susceptibility to dextran sulfate sodium-induced colitis. J. Immunol. 2013; 191:325-2143. doi: 10.4049/jimmunol.1203363. PMID: 23913970
Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, Dunaway DL, Fell HP, Ferree S, George RD, Grogan T, James JJ, Maysuria M, Mitton JD, Oliveri P, Osborn JL, Peng T, Ratcliffe AL, Webster PJ, Davidson EH, Hood L, Dimitrov K. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol. 2008 Mar;26(3):317-25. doi: 10.1038/nbt1385. Epub 2008 Feb 17. PMID:18278033
Sapkota B, Hill CE, Pollack BP. Vemurafenib enhances MHC induction in BRAF(V600E) homozygous melanoma cells. Oncoimmunology. 2013 Jan 1;2(1):e22890. PubMed PMID: 23483066; PubMed Central PMCID: PMC3583938.
Ullal AV, Weissleder R. Photocleavable DNA Barcoding Antibodies for Multiplexed Protein Analysis in Single Cells. Methods Mol Biol. 2015;1346:47-54. doi: 10.1007/978-1-4939-2987-0_4. PMID: 26542714
Wang H, Lee S, Nigro CL, Lattanzio L, Merlano M, Monteverde M, Matin R, Purdie K, Mladkova N, Bergamaschi D, Harwood C, Syed N, Szlosarek P, Briasoulis E, McHugh A, Thompson A, Evans A, Leigh I, Fleming C, Inman GJ, Hatzimichael E, Proby C, Crook T. NT5E (CD73) is epigenetically regulated in malignant melanoma and associated with metastatic site specificity. Br J Cancer. 2012 Apr 10;106(8):1446-52. doi: 10.1038/bjc.2012.95. Epub 2012 Mar 27. PubMed PMID: 22454080; PubMed Central PMCID: PMC3326678.
Joseph M. Beechem, Ph.D. ([email protected]), is senior vice president of research and development and Sarah E. Warren, Ph.D. ([email protected]), is senior scientist, immune oncology at NanoString.







