Microarray technology has become a standard tool for gene-expression analysis. To generate reliable microarray data that can be compared between different experiments and laboratories, however, the target RNA must be of high quality and reproducibility. Traditional methods of target RNA preparation have been time consuming and error prone. With the largely manual techniques, results vary from technician to technician. Pipetting errors also result in costly reagent waste.
Beckman Coulter (www.beckmancoulter.com)has developed an automated target preparation solution that reduces variability from human error and saves time. The development of this application was based on our earlier application on the Biomek FXP workstation.
The ArrayPlex application on the Biomek 3000 workstation integrates the liquid handler, a Peltier static device, a Peltier shaker, and an optional customer- supplied thermocycler. The highly automated process includes three methods: cDNA synthesis, in vitro transcription, and fragmentation. A full or partial 96-well plate of samples can be processed in multiples of eight in a single run.
In the example presented here, Affymetrix’ (www.affymetrix.com) GeneChip Expression 3´-Amplification reagents were used for cDNA synthesis and cRNA amplification and labeling. The Agencourt RNAClean kit, based on the patented SPRI® (solid-phase reversible immobilization) paramagnetic bead-based technology, was used during cDNA and cRNA cleanup.
In this study, eight plates of 24 samples of 1 µg and 2 µg of human HeLa total RNA (Ambion) were processed using the automated methods. Forty-eight of the amplified cRNA samples were randomly selected to hybridize to the Affymetrix Human Genome U133 Plus 2.0 Array.
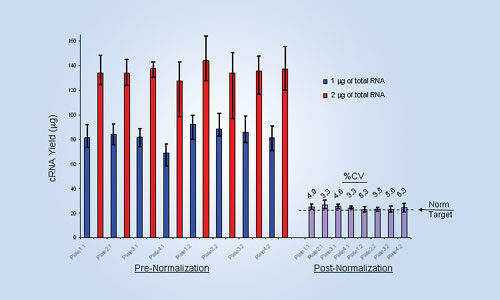
Figure 1
Methods
The automated methods process a full or partial 96-well plate of samples in a single run. A screen prompt at system start allows the users to enter the number of samples to run. The first two methods are comprised of two independent steps: cDNA synthesis/ cDNA cleanup; cRNA synthesis/cRNA cleanup. The third method is comprised of three independent steps: quantitation, normalization, and fragmentation. Users can save samples after cDNA synthesis, cRNA synthesis, or quantitation and run the remaining processing steps at a later time. The decks must be refreshed before cDNA and cRNA cleanup.
Eight plates of 24 samples were processed using the ArrayPlex applications on the Biomek 3000 workstation. Human HeLa total RNA was used as the starting material. Twenty-four 1 µg of total RNA and 24 2 µg of total RNA were randomly distributed in each column. The cRNA concentration was measured using the OD260 readings, and the cRNA yield was calculated based on a 35 µL final volume.
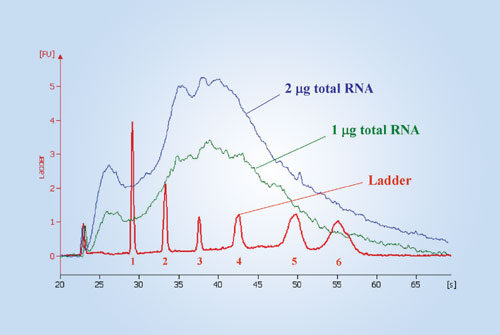
Figure 2
Results
The average cRNA yield (Figure 1) from 1 µg of total RNA (shown in blue) was 83.1 µg. The average cRNA yield from 2 µg of total RNA (shown in red) was 135.6 µg for plate 1 and 104.4 µg for Plate 2. The normalization target was 21.8 µg, and the post normalization CVs were <10%. The error bars show the highest and lowest yields in each group.
cRNA samples were analyzed on the Agilent 2100 Bioanalyzer (Figure 2). Ambion’s RNA 6000 Ladder was used as the marker. The peak sizes are 0.2, 0.5, 1.0, 2.0, 4.0, and 6.0 kb for peaks one through six. Electropherograms showed similar nondegraded cRNA profiles with cRNA samples amplified from 1 µg and 2 µg of total RNA.
A total of 48 cRNA samples were hybridized to the Affymetrix Human Genome U133 Plus 2.0 Array. Samples were run on two different Biomek 3000 ArrayPlex instruments by two different technicians and were hybridized using multiple lots of GeneChips on different days. The data showed good correlation and reproducibility between samples.
Log correlation comparison analysis of samples with the varying amount of total RNA was prepared using GCOS 1.3 (Figure 3). The blue diagonal bars indicate a twofold change. The correlation comparison of two samples within the blue diagonal bars indicates good correlation from sample to sample, plate to plate, and sample concentration to sample concentration.
This study shows that automation can deliver consistent plate-to-plate, sample-to-sample yields. Confirming 2100 Bioanalyzer electropherograms showed similar nondegraded profiles for cRNA generated from 1 and 2 µg of total RNA, indicating no performance bias with different amounts of starting materials. The highly automated steps reduce errors, reagent waste, and deliver reproducible results.
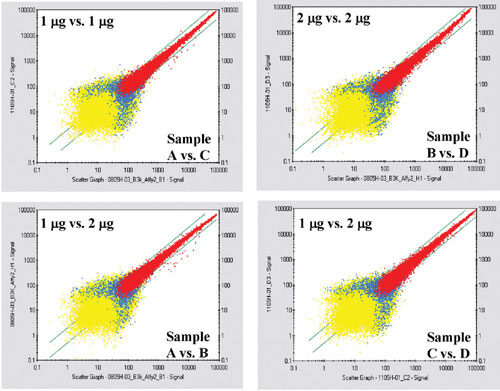
Figure 3
Zhu Zhu is senior applications scientist, Handy Yowanto, Ph.D., is staff applications scientist, and Keith Roby is product manager, automation information systems, at Beckman Coulter. Web: www.beckmancoulter.com. Phone: (714) 961-4228.
E-mail: [email protected].







