October 1, 2007 (Vol. 27, No. 17)
New Tool Provides Greater Multiplexing with Reproducible Data Quality
A new wave of automated, inexpensive, and high-throughput multiplexing technologies has been widely reported on of late. Diagnostic laboratories and clinical pharmacologists will find these multiplexing systems ideal for identifying and validating biomarkers for diverse applications ranging from testing for adverse drug reactions (ADR) to prognostics and diagnostics. In this tutorial, we discuss the application of one of these multiplex technologies for validating DNA biomarkers.
The use of highly multiplexed genotyping arrays to interrogate several hundred thousand single nucleotide polymorphisms (SNPs) per sample has made possible significant leaps in the understanding of the human genome.
Such genome-scale solutions are particularly effective for initial discovery work where the goal is to begin the process of correlating gene mutations with disease phenotypes. This is achieved by comparing SNPs across hundreds to thousands of samples obtained from both healthy and diseased sources.
Since there are more SNPs in the human genome than is currently possible to assay on one chip, Illumina (www.illumina.com) uses tag SNPs representing specific regions of the genome rather than one individual mutation. The initial discovery work using highly multiplexed arrays may identify up to several hundred tag SNPs.
To identify the precise biomarkers associated with disease, researchers must map DNA regions and characterize many more SNPs per region. At this stage, there are far fewer targets analyzed per sample, but more samples are analyzed overall. Because of the focused analysis on fewer targets per sample, highly multiplexed systems are no longer cost-effective, and the cost emphasis changes from striving for the lowest cost per SNP to seeking the lowest cost per sample.
Conversely, single-plex technologies may be quick and inexpensive for individual targets but become too costly when applied to many targets per sample. Thus there is a growing need for technologies like Illumina’s VeraCode™, which can analyze one to several hundred targets per sample.
Biomarker validation is the confirmatory step in determining whether a particular mutation is associated with a disease. At this point, thousands of samples from both healthy and diseased sources are screened for the relevant biomarkers. In addition to the constraints mentioned previously, the data generated during biomarker validation must be reliable and reproducible if SNPs are to be used later to aid in the diagnosis of a particular disease or for guiding prognosis of disease outcome.
Illumina’s VeraCode technology specifically addresses the need for a low- to mid-level multiplexing, high throughput, and low cost-per-sample instrument capable of processing the focused assays necessary for biomarker validation.
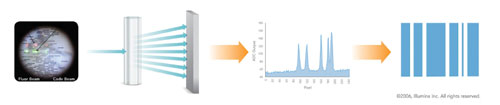
At the heart of VeraCode technology are cylindrical glass beads measuring 240 microns in length by 28 microns in diameter. Each VeraCode microbead is inscribed with a unique digital holographic code to designate and track the specific analyte or genotype of interest (Figure).
Microbeads
The unique code enables use of the microbeads for tracking critical identifiers such as sample ID, laboratory ID, and reagent kits in addition to the multiplex assay markers. Up to 24 bits of information inscribed in each code allows for a high level of error checking, improving the robustness of the optical readout process and providing a high level of reliability. Unlike traditional microarrays, VeraCode microbeads take advantage of solution-phase kinetics for more rapid hybridization times that dramatically reduce time to results.
Removing the Guesswork from ID
Assays developed for the VeraCode microbeads are analyzed with Illumina’s BeadXpress Reader, a high-throughput, two-color laser detection system designed to be compatible with standard laboratory automation. This system uses both red and green lasers to measure two fluorescent signatures from each bead and enable the processing of two-color assays. Additionally, the green laser is split off and used to interrogate the embedded digital holographic element and classify the beads.
In two-color genotyping assays, each bead type represents a different locus, and each fluorescent probe represents one allele that can be present at that locus. A bead with one fluorescent signal (red or green) would be homozygous for one of the alleles, and a bead displaying both fluorescent signals would be heterozygous.
This two-color detection system has several advantages over competitive single-color detection systems. Until now, comparable technologies have suffered from two major setbacks—limited multiplexing ability and inconsistent data quality from one assay application to another. VeraCode’s holographic codes and bead classification process have been designed to overcome these limitations.
Greater Multiplexing
Earlier generations of flow cytometry-based systems require fluorescent signatures to classify bead types, thus they are limited to single-color detection assays. This means that for every locus in a genotyping assay, two bead types must be used to generate a genotyping call.
In contrast, the BeadXpress Reader uses a holographic code in order to classify the bead types, thus both fluorescent detection channels are free to be used in the assay itself. As such, one can deploy the two-color GoldenGate® Genotyping Assay on the BeadXpress, which allows users to utilize one bead type per locus in order to make a genotyping call. Alternatively, BeadXpress offers users the flexibility to use single-color detection assays if they prefer this methodology.
Golden Data Quality
The BeadXpress Reader uses a green laser to read the embedded holographic element contained within each bead. This element contains up to a 24 bit digital code with internal error checking to prevent misclassification.
A necessary concern for any researcher considering new genotyping technology is the assay’s reliability. VeraCode uses the GoldenGate Genotyping Assay, which is the same assay researchers used with Illumina’s BeadArray technology to generate the data for completion of 70% of the Phase I International HapMap Project. To date, more than one million SNP assays have been developed using GoldenGate technology.
The BeadXpress reader allows researchers to thoroughly analyze data. Each unique VeraCode bead type used to represent a particular biomarker is represented with an average redundancy of 15–22 beads, with the data for each individual bead made available for analysis.
Furthermore, the fluorescence intensities for each bead are sampled up to 12 times per bead providing highly robust data. Although this provides significantly more data than is needed to make successful genotyping calls, the availability of this data allows researchers to calculate inter- and intrabead coefficient of variants, which is critical in assay validation and quality metrics.
The Future of Diagnostic Testing
Systems such as BeadXpress have opened exciting new possibilities in molecular diagnostic testing. The Children’s Hospital of Eastern Ontario is currently using the VeraCode technology to develop diagnostic tests for newborn screening. Mayo Clinic has also entered into a diagnostic collaborative agreement to codevelop molecular diagnostic tests for the study of complex diseases.
Other applications such as patient screening for ADR may soon be within reach thanks to high-throughput, mid-plex genotyping systems. With the ability to tie specific biomarkers to ADR, pharmaceutical companies would be able to safely market and sell high-efficacy drugs that would otherwise be grounded due to deadly side effects in small pockets of the population.
Mickie Henshall is marketing manager, molecular diagnostics, and Elliott Gorfain is product manager, VeraCode™ technology, at Illumina. Web: www.illumina.com.
Phone: (858) 202-4551.
E-mail: [email protected].



