DNA and RNA are the key information-rich molecules in cells, comprising nearly a quarter of the dry weight of E. coli, for example. Methods for extracting and purifying nucleic acids play a central role in contemporary biosciences because nucleic acids are intimate participants in the storage and retrieval of hereditary information.
Extraction procedures lyse cells and prepare nucleic acids for subsequent analysis, such as end-point PCR, which does not require highly pure starting material. Purification procedures, in contrast, remove proteins, lipids, carbohydrates, and cell debris, leaving concentrated nucleic acids suitable for cloning, sequencing, Northern or Southern blotting, RT-PCR, and microarray analysis. Because purification precedes further often complex downstream analysis, it is essential to invest some thought and effort into one’s purification methods and reagents.
The three most common purification technologies are magnetic beads or particles, spin columns with disks that adsorb nucleic acids, and selective precipitation. A few advantages and disadvantages of these methods are summarized in the Table.
Spin columns are the most commonly used technology, although for robotic high-throughput applications, magnetic beads are easily adaptable to and are popular for 96-well plate formats. On the other hand, the total yield from spin columns is lower than for selective precipitation, and high molecular weight molecules are not efficiently released from spin column adsorbent disks. Selective precipitation requires more centrifugation steps but delivers higher yields.
The purification technology chosen depends ultimately on the needs of the user. If the goal is to obtain some nucleic acid from dozens or hundreds of samples, for example, then a rapid or high-throughput procedure is warranted. If, on the other hand, high molecular weight nucleic acids are desired, then it would be wise to use a slower, but more robust method for retaining larger size classes of molecules.
Environmental Samples
Extraction of nucleic acids from microbial cells in environmental samples, such as soils, sediments and surface waters, poses unique challenges. For example, it may be difficult to separate cellular material from the physical matrix. Some soil DNA extraction methods utilize beads of various sizes to help break down the matrix and release cells from it. Unfortunately, this “bead-beating,” as it is called, also sheers DNA molecules, reducing the longer strands to shorter ones. DNA isolated by the SoilMaster™ DNA Extraction Kit from Epicentre Biotechnologies (www.epibio.com) contains much larger DNA molecules than paired samples prepared by bead-beating methods.
This difference in the sizes of DNA obtained was confirmed by field inversion gel electrophoresis (data not shown). The yield of DNA from SoilMaster is also greater, even though the bead-beating method was applied to an equivalent amount of soil. Another bothersome attribute of soil matrices is the presence of humic acids which inhibit PCR. To address this drawback, SoilMaster includes a simple method for removal of inhibitory humic acids.
Scarce Source Material
Retrieval of nucleic acids from a few hundred eukaryotic cells, such as those obtained from laser-capture methods, is another purification challenge. To highlight the capabilities of the selective precipitation method in this regard, HeLa cells were aseptically diluted, trapped inside sterile 5-µL microcapillary pipettes, and counted using an inverted microscope.
After elution, the cells were washed with phosphate-buffered saline, RNA was purified using the Epicentre ArrayPure™ Nano-scale RNA Purification Kit, the RNA was reverse-transcribed with Epicentre’s MMLV Reverse Transcriptase, and the resulting cDNA was quantified by qPCR.

Figure 1A shows the amplification plots detecting cDNA corresponding to RNA purified from 1, 10, and 100 cells and from the media control (0 cells). Figure 1B is a standard curve indicating that over three orders of magnitude there is a linear relationship between log of initial cell number and cycle threshold.
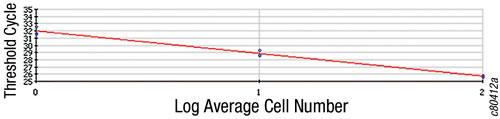
The RNA from 20 HeLa cells (400-pg total RNA) has successfully been amplified in a two-round RNA amplification procedure using the Epicentre TargetAmp™ 2-Round aRNA Amplification Kit 2.0. A 32-µg amount of amplified RNA was produced after two rounds of amplification, and the amplified RNA could be used in RT-PCR. Thus, it is possible to quantitatively purify RNA from a few cells by means of the selective precipitation method and then amplify that RNA.
Conclusion
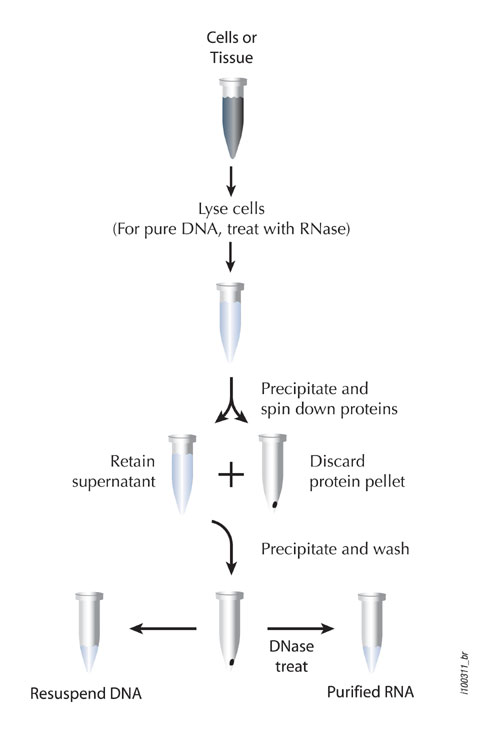
An overview of selective precipitation is found in Figure 2. The chief advantages of this technology are the higher yields obtained and the retention of high molecular weight species of nucleic acids.
Bruce W. Jarvis, Ph.D., is purification product manager, and Haiying Grunenwald, Ph.D., is senior R&D scientist at Epicentre Biotechnologies. Phone: (608) 442-6116. E-mail: [email protected].